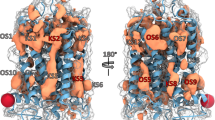
Abstract
Membrane receptors sense and transduce extracellular stimuli into intracellular signaling responses but the molecular underpinnings remain poorly understood. We report a computational approach for designing protein allosteric signaling functions. By combining molecular dynamics simulations and design calculations, the method engineers amino-acid ‘microswitches’ at allosteric sites that modulate receptor stability or long-range coupling, to reprogram specific signaling properties. We designed 36 dopamine D2 receptor variants, whose constitutive and ligand-induced signaling agreed well with our predictions, repurposed the D2 receptor into a serotonin biosensor and predicted the signaling effects of more than 100 known G-protein-coupled receptor (GPCR) mutations. Our results reveal the existence of distinct classes of allosteric microswitches and pathways that define an unforeseen molecular mechanism of regulation and evolution of GPCR signaling. Our approach enables the rational design of allosteric receptors with enhanced stability and function to facilitate structural characterization, and reprogram cellular signaling in synthetic biology and cell engineering applications.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Code availability
Protein modeling and design applications (IPHoLD, RosettaMembrane) are available in the latest version of the Rosetta software package (https://www.rosettacommons.org/software) or at https://doi.org/10.5281/zenodo.3460811. Bio3D is available at: http://thegrantlab.org/bio3d/index.php.
References
Suel, G. M., Lockless, S. W., Wall, M. A. & Ranganathan, R. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat. Struct. Biol. 10, 59–69 (2003).
Reynolds, K. A., McLaughlin, R. N. & Ranganathan, R. Hot spots for allosteric regulation on protein surfaces. Cell 147, 1564–1575 (2011).
Monod, J., Wyman, J. & Changeux, J. P. On the nature of allosteric transitions: a plausible model. J. Mol. Biol. 12, 88–118 (1965).
Cui, Q. & Karplus, M. Allostery and cooperativity revisited. Protein Sci. 17, 1295–1307 (2008).
Luque, I., Leavitt, S. A. & Freire, E. The linkage between protein folding and functional cooperativity: two sides of the same coin? Annu Rev. Biophys. Biomol. Struct. 31, 235–256 (2002).
Clarkson, M. W., Gilmore, S. A., Edgell, M. H. & Lee, A. L. Dynamic coupling and allosteric behavior in a nonallosteric protein. Biochemistry 45, 7693–7699 (2006).
Bhattacharya, S. & Vaidehi, N. Differences in allosteric communication pipelines in the inactive and active states of a GPCR. Biophys. J. 107, 422–434 (2014).
Miao, Y., Nichols, S. E., Gasper, P. M., Metzger, V. T. & McCammon, J. A. Activation and dynamic network of the M2 muscarinic receptor. Proc. Natl Acad. Sci. USA 110, 10982–10987 (2013).
Kohlhoff, K. J. et al. Cloud-based simulations on google exacycle reveal ligand modulation of GPCR activation pathways. Nat. Chem. 6, 15–21 (2014).
McLaughlin, R. N. Jr., Poelwijk, F. J., Raman, A., Gosal, W. S. & Ranganathan, R. The spatial architecture of protein function and adaptation. Nature 491, 138–142 (2012).
Sung, Y. M., Wilkins, A. D., Rodriguez, G. J., Wensel, T. G. & Lichtarge, O. Intramolecular allosteric communication in dopamine D2 receptor revealed by evolutionary amino acid covariation. Proc. Natl Acad. Sci. USA 113, 3539–3544 (2016).
Rosenbaum, D. M., Rasmussen, S. G. & Kobilka, B. K. The structure and function of G-protein-coupled receptors. Nature 459, 356–363 (2009).
Tsai, C. J. & Nussinov, R. A unified view of ‘how allostery works’. PLoS Comput. Biol. 10, e1003394 (2014).
Motlagh, H. N., Wrabl, J. O., Li, J. & Hilser, V. J. The ensemble nature of allostery. Nature 508, 331–339 (2014).
Wisler, J. W., Xiao, K., Thomsen, A. R. & Lefkowitz, R. J. Recent developments in biased agonism. Curr. Opin. Cell Biol. 27, 18–24 (2014).
Venkatakrishnan, A. J. et al. Molecular signatures of G-protein-coupled receptors. Nature 494, 185–194 (2013).
Dror, R. O. et al. Activation mechanism of the beta2-adrenergic receptor. Proc. Natl Acad. Sci. USA 108, 18684–18689 (2011).
Ahuja, S. & Smith, S. O. Multiple switches in G protein-coupled receptor activation. Trends Pharm. Sci. 30, 494–502 (2009).
Katritch, V., Cherezov, V. & Stevens, R. C. Diversity and modularity of G protein-coupled receptor structures. Trends Pharm. Sci. 33, 17–27 (2012).
Flock, T. et al. Selectivity determinants of GPCR-G-protein binding. Nature 545, 317–322 (2017).
Scott, D. J., Kummer, L., Tremmel, D. & Pluckthun, A. Stabilizing membrane proteins through protein engineering. Curr. Opin. Chem. Biol. 17, 427–435 (2013).
Magnani, F., Shibata, Y., Serrano-Vega, M. J. & Tate, C. G. Co-evolving stability and conformational homogeneity of the human adenosine A2a receptor. Proc. Natl Acad. Sci. USA 105, 10744–10749 (2008).
Egloff, P. et al. Structure of signaling-competent neurotensin receptor 1 obtained by directed evolution in Escherichia coli. Proc. Natl Acad. Sci. USA 111, E655–E662 (2014).
Sarkar, C. A. et al. Directed evolution of a G protein-coupled receptor for expression, stability, and binding selectivity. Proc. Natl Acad. Sci. USA 105, 14808–14813 (2008).
Ye, L., Van Eps, N., Zimmer, M., Ernst, O. P. & Prosser, R. S. Activation of the A2A adenosine G-protein-coupled receptor by conformational selection. Nature 533, 265–268 (2016).
LeVine, M. V. & Weinstein, H. AIM for allostery: using the Ising model to understand information processing and transmission in allosteric biomolecular systems. Entropy 17, 2895–2918 (2015).
Feng, X., Ambia, J., Chen, K. M., Young, M. & Barth, P. Computational design of ligand-binding membrane receptors with high selectivity. Nat. Chem. Biol. 13, 715–723 (2017).
Chen, K. Y., Sun, J., Salvo, J. S., Baker, D. & Barth, P. High-resolution modeling of transmembrane helical protein structures from distant homologues. PLoS Comput. Biol. 10, e1003636 (2014).
Barth, P., Schonbrun, J. & Baker, D. Toward high-resolution prediction and design of transmembrane helical protein structures. Proc. Natl Acad. Sci. USA 104, 15682–15687 (2007).
Chen, K. Y., Zhou, F., Fryszczyn, B. G. & Barth, P. Naturally evolved G protein-coupled receptors adopt metastable conformations. Proc. Natl Acad. Sci. USA 109, 13284–13289 (2012).
Perica, T. et al. Evolution of oligomeric state through allosteric pathways that mimic ligand binding. Science 346, 1254346 (2014).
Bahar, I., Lezon, T. R., Yang, L. W. & Eyal, E. Global dynamics of proteins: bridging between structure and function. Annu. Rev. Biophys. 39, 23–42 (2010).
J Luo, Y Zhu, M Zhu, H Hu, 2011 Cell-based calcium assay for medium to high throughput screening of TRP channel functions using FlexStation 3. J. Vis. Exp. 17.
Ballesteros, J. A. & Weinstein, H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 25, 366–428 (1995).
Kang, Y. et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature 523, 561–567 (2015).
Rasmussen, S. G. et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature 477, 549–555 (2011).
Koehl, A. et al. Structure of the micro-opioid receptor-Gi protein complex. Nature 558, 547–552 (2018).
Carpenter, B., Nehme, R., Warne, T., Leslie, A. G. & Tate, C. G. Structure of the adenosine A(2A) receptor bound to an engineered G protein. Nature 536, 104–107 (2016).
Yao, X. J. et al. The effect of ligand efficacy on the formation and stability of a GPCR-G protein complex. Proc. Natl Acad. Sci. USA 106, 9501–9506 (2009).
Isberg, V. et al. GPCRdb: an information system for G protein-coupled receptors. Nucleic Acids Res. 45, 2936 (2017).
Kellogg, E. H., Leaver-Fay, A. & Baker, D. Role of conformational sampling in computing mutation-induced changes in protein structure and stability. Proteins 79, 830–838 (2011).
Rodriguez, G. J., Yao, R., Lichtarge, O. & Wensel, T. G. Evolution-guided discovery and recoding of allosteric pathway specificity determinants in psychoactive bioamine receptors. Proc. Natl Acad. Sci. USA 107, 7787–7792 (2010).
Schonegge, A. M. et al. Evolutionary action and structural basis of the allosteric switch controlling beta2AR functional selectivity. Nat. Commun. 8, 2169 (2017).
Han, M., Smith, S. O. & Sakmar, T. P. Constitutive activation of opsin by mutation of methionine 257 on transmembrane helix 6. Biochemistry 37, 8253–8261 (1998).
Graber, S. G., Figler, R. A. & Garrison, J. C. Expression and purification of functional G protein alpha subunits using a baculovirus expression system. J. Biol. Chem. 267, 1271–1278 (1992).
Miller, M. et al. in TRP Channels (ed. Zhu, M. X.) 1–20 (Boca Raton, FL, CRC Press/Taylor & Francis, 2011).
Young, M. et al. Computational design of orthogonal membrane receptor-effector switches for rewiring signaling pathways. Proc. Natl Acad. Sci. USA 115, 7051–7056 (2018).
Davis, I. W. & Baker, D. RosettaLigand docking with full ligand and receptor flexibility. J. Mol. Biol. 385, 381–392 (2009).
Befort, K., Zilliox, C., Filliol, D., Yue, S. & Kieffer, B. L. Constitutive activation of the delta opioid receptor by mutations in transmembrane domains III and VII. J. Biol. Chem. 274, 18574–18581 (1999).
Cavalli, A., Babey, A. M. & Loh, H. H. Altered adenylyl cyclase responsiveness subsequent to point mutations of Asp 128 in the third transmembrane domain of the delta-opioid receptor. Neuroscience 93, 1025–1031 (1999).
Decaillot, F. M. et al. Opioid receptor random mutagenesis reveals a mechanism for G protein-coupled receptor activation. Nat. Struct. Biol. 10, 629–636 (2003).
Han, M., Lin, S. W., Minkova, M., Smith, S. O. & Sakmar, T. P. Functional interaction of transmembrane helices 3 and 6 in rhodopsin. Replacement of phenylalanine 261 by alanine causes reversion of phenotype of a glycine 121 replacement mutant. J. Biol. Chem. 271, 32337–32342 (1996).
Han, M., Lin, S. W., Smith, S. O. & Sakmar, T. P. The effects of amino acid replacements of glycine 121 on transmembrane helix 3 of rhodopsin. J. Biol. Chem. 271, 32330–32336 (1996).
Acknowledgements
We thank R. Sharma for setting up some NMA calculations, T. Wensel and his laboratory for sharing experimental protocols and members of the Barth laboratory for helpful discussion. This work was supported by funding from EPFL and the Ludwig Institute for Cancer Research, partially supported by a grant from the National Institute of Health (no. 1R01GM097207), and by a supercomputer allocation from XSEDE (no. MCB120101) to P.B.
Author information
Authors and Affiliations
Contributions
P.B. designed the studies. P.B., K.-Y.M.C. and D.K. performed the calculations. K.-Y.M.C. and D.K. performed experiments on D2. P.B., D.K. and K.-Y.M.C. analyzed the data. P.B. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
P.B. declares the following patent application: patent applicant, Ecole Polytechnique Fédérale de Lausanne. Name of inventor(s), Patrick Barth. Application number, EP 19189259.5. Status of application, priority year. Specific aspect of manuscript covered in patent application, methods and protein variants.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–5, Supplementary Figs. 1–11 and Supplementary Note.
Rights and permissions
About this article
Cite this article
Chen, KY.M., Keri, D. & Barth, P. Computational design of G Protein-Coupled Receptor allosteric signal transductions. Nat Chem Biol 16, 77–86 (2020). https://doi.org/10.1038/s41589-019-0407-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-019-0407-2
This article is cited by
-
G protein-coupled receptors (GPCRs): advances in structures, mechanisms, and drug discovery
Signal Transduction and Targeted Therapy (2024)
-
A Vaccinia-based system for directed evolution of GPCRs in mammalian cells
Nature Communications (2023)
-
Differential sensing with arrays of de novo designed peptide assemblies
Nature Communications (2023)
-
Prediction of dynamic allostery for the transmembrane domain of the sweet taste receptor subunit, TAS1R3
Communications Biology (2023)
-
Computational design of dynamic receptor—peptide signaling complexes applied to chemotaxis
Nature Communications (2023)