Abstract
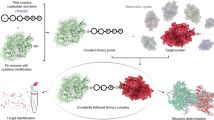
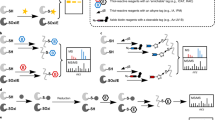
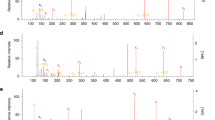
Itaconate has been recently recognized as an anti-inflammatory metabolite involved in the pathogen–macrophage interface. Due to its weak electrophilicity, itaconate could modify cysteines of the protein KEAP1 and glutathione, which contribute to its anti-inflammatory effect. However, the substrates of itaconate modification in macrophages have not been systematically profiled, which largely impedes the understanding of its roles in immune responses. Here, we developed a specific thiol-reactive probe, 1-OH-Az, for quantitative chemoproteomic profiling of cysteine modifications by itaconate, and provided a global portrait of its proteome reactivity. We found that itaconate covalently modifies key glycolytic enzymes and impairs glycolytic flux mainly through inhibition of fructose-bisphosphate aldolase A (ALDOA). Moreover, itaconate attenuates the inflammatory response in stimulated macrophages by impairing the glycolysis. Our study provides a valuable resource of protein targets of itaconate in macrophages and establishes a negative-feedback link between glycolysis and itaconate, elucidating new functional insights for this anti-inflammatory metabolite.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The proteomics data (ID: 206029, 206030, 206031) has been deposited at https://chorusproject.org/pages/index.html. The data that support the findings of this study are available from the corresponding authors upon request.
References
Kelly, B. & O’Neill, L. A. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 25, 771–784 (2015).
Vats, D. et al. Oxidative metabolism and PGC-1beta attenuate macrophage-mediated inflammation. Cell Metab. 4, 13–24 (2006).
Palsson-McDermott, E. M. et al. Pyruvate kinase M2 regulates Hif-1α activity and IL-1β induction and is a critical determinant of the Warburg effect in LPS-activated macrophages. Cell Metab. 21, 65–80 (2015).
Van den Bossche, J., O’Neill, L. A. & Menon, D. Macrophage immunometabolism: where are we (going)? Trends Immun. 38, 395–406 (2017).
Kornberg, M. D. et al. Dimethyl fumarate targets GAPDH and aerobic glycolysis to modulate immunity. Science 360, 449–453 (2018).
Tannahill, G. M. et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 496, 238–242 (2013).
Murphy, M. P. & O’Neill, L. A. J. Krebs cycle reimagined: the emerging roles of succinate and itaconate as signal transducers. Cell 174, 780–784 (2018).
Lampropoulou, V. et al. Itaconate links inhibition of succinate dehydrogenase with macrophage metabolic remodeling and regulation of inflammation. Cell Metab. 24, 158–166 (2016).
Dominguez-Andres, J. et al. The itaconate pathway is a central regulatory node linking innate immune tolerance and trained immunity. Cell Metab. 29, 211–220 (2019).
Michelucci, A. et al. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc. Natl Acad. Sci. USA 110, 7820–7825 (2013).
Strelko, C. L. et al. Itaconic acid is a mammalian metabolite induced during macrophage activation. J. Am. Chem. Soc. 133, 16386–16389 (2011).
Sasikaran, J., Ziemski, M., Zadora, P. K., Fleig, A. & Berg, I. A. Bacterial itaconate degradation promotes pathogenicity. Nat. Chem. Biol. 10, 371–377 (2014).
Shen, H. et al. The human knockout gene CLYBL connects itaconate to vitamin B12. Cell 171, 771–782 (2017).
O’Neill, L. A. J. & Artyomov, M. N. Itaconate: the poster child of metabolic reprogramming in macrophage function. Nat. Rev. Immunol. 19, 273–281 (2019).
Mills, E. L. et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 556, 113–117 (2018).
Bambouskova, M. et al. Electrophilic properties of itaconate and derivatives regulate the IκBζ-ATF3 inflammatory axis. Nature 556, 501–504 (2018).
Nomura, D. K., Dix, M. M. & Cravatt, B. F. Activity-based protein profiling for biochemical pathway discovery in cancer. Nat. Rev. Cancer 10, 630–638 (2010).
Niphakis, M. J. & Cravatt, B. F. Enzyme inhibitor discovery by activity-based protein profiling. Ann. Rev. Biochem. 83, 341–377 (2014).
Weerapana, E. et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468, 790–U779 (2010).
Shannon, D. A. et al. Investigating the proteome reactivity and selectivity of Aryl halides. J. Am. Chem. Soc. 136, 3330–3333 (2014).
Abegg, D. et al. Proteome-Wide profiling of targets of cysteine reactive small molecules by using ethynyl benziodoxolone reagents. Angew. Chem. Int. Edit. 54, 10852–10857 (2015).
Akter, S. et al. Chemical proteomics reveals new targets of cysteine sulfinic acid reductase. Nat. Chem. Biol. 14, 995–1004 (2018).
Hacker, S. M. et al. Global profiling of lysine reactivity and ligandability in the human proteome. Nat. Chem. 9, 1181–1190 (2017).
Yang, J., Gupta, V., Carroll, K. S. & Liebler, D. C. Site-specific mapping and quantification of protein S-sulphenylation in cells. Nat. Commun. 5, 4776 (2014).
Gupta, V., Yang, J., Liebler, D. C. & Carroll, K. S. Diverse redoxome reactivity profiles of carbon nucleophiles. J. Am. Chem. Soc. 139, 5588–5595 (2017).
Lentz, C. S. et al. Identification of a S. aureus virulence factor by activity-based protein profiling (ABPP). Nat. Chem. Biol. 14, 609–617 (2018).
Wang, C., Weerapana, E., Blewett, M. M. & Cravatt, B. F. A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles. Nat. Methods 11, 79–85 (2014).
Kulkarni, R. A. et al. A chemoproteomic portrait of the oncometabolite fumarate. Nat. Chem. Biol. 15, 391–400 (2019).
Counihan, J. L., Wiggenhorn, A. L., Anderson, K. E. & Nomura, D. K. Chemoproteomics-enabled covalent ligand screening reveals ALDH3A1 as a lung cancer therapy target. ACS Chem. Biol. 13, 1970–1977 (2018).
Tian, C. et al. Multiplexed thiol reactivity profiling for target discovery of electrophilic natural products. Cell Chem. Biol. 24, 1416–1427 (2017).
Blewett, M. M. et al. Chemical proteomic map of dimethyl fumarate-sensitive cysteines in primary human T cells. Sci. Signal 9, 445 (2016).
Backus, K. M. et al. Proteome-wide covalent ligand discovery in native biological systems. Nature 534, 570–574 (2016).
Qin, W. et al. Artificial cysteine S-glycosylation induced by per-O-acetylated unnatural monosaccharides during metabolic glycan labeling. Angew. Chem. Int. Edn 57, 1817–1820 (2018).
Weerapana, E., Simon, G. M. & Cravatt, B. F. Disparate proteome reactivity profiles of carbon electrophiles. Nat. Chem. Bio. 4, 405–407 (2008).
Kolb, H. C., Finn, M. G. & Sharpless, K. B. Click chemistry: diverse chemical function from a few good reactions. Angew. Chem. Int. Edn 40, 2004–2021 (2001).
Qin, K. et al. Quantitative profiling of protein O-GlcNAcylation sites by an isotope-tagged cleavable linker. ACS Chem. Bio. 13, 1983–1989 (2018).
O’Shea, J. P. et al. pLogo: a probabilistic approach to visualizing sequence motifs. Nat. Methods 10, 1211–1212 (2013).
Yao, D. C. et al. Hemolytic anemia and severe rhabdomyolysis caused by compound heterozygous mutations of the gene for erythrocyte/muscle isozyme of aldolase, ALDOA((Arg303X/Cys338Tyr)). Blood 103, 2401–2403 (2004).
Jang, C., Chen, L. & Rabinowitz, J. D. Metabolomics and isotope tracing. Cell 173, 822–837 (2018).
Fu, J. et al. Hyperactivity of the transcription factor Nrf2 causes metabolic reprogramming in mouse esophagus. J. Bio. Chem. 294, 327–340 (2018).
Singh, A. et al. Small molecule inhibitor of NRF2 selectively intervenes therapeutic resistance in KEAP1-Deficient NSCLC tumors. ACS Chem. Biol. 11, 3214–3225 (2016).
Puchalska, P. et al. Isotope tracing untargeted metabolomics reveals macrophage polarization-state-specific metabolic coordination across intracellular compartments. iScience 9, 298–313 (2018).
Qin, W. et al. Quantitative time-resolved chemoproteomics reveals that stable O-GlcNAc regulates box C/D snoRNP biogenesis. Proc. Natl Acad. Sci. USA 114, E6749–E6758 (2017).
Tarbet, H. J., Toleman, C. A. & Boyce, M. A sweet embrace: control of protein-protein interactions by O-Linked beta-N-Acetylglucosamine. Biochemistry 57, 13–21 (2018).
Chen, Y. et al. Quantitative profiling of protein carbonylations in ferroptosis by an aniline-derived probe. J. Am. Chem. Soc. 140, 4712–4720 (2018).
Yang, J., Tallman, K. A., Porter, N. A. & Liebler, D. C. Quantitative chemoproteomics for site-specific analysis of protein alkylation by 4-hydroxy-2-nonenal in cells. Anal. Chem. 87, 2535–2541 (2015).
Isobe, Y. et al. Identification of protein targets of 12/15-Lipoxygenase-derived lipid electrophiles in mouse peritoneal macrophages using omega-alkynyl fatty acid. ACS Chem. Biol. 13, 887–893 (2018).
Moellering, R. E. & Cravatt, B. F. Functional lysine modification by an intrinsically reactive primary glycolytic metabolite. Science 341, 549–553 (2013).
Bollong, M. J. et al. A metabolite-derived protein modification integrates glycolysis with KEAP1-NRF2 signalling. Nature 562, 600–604 (2018).
Huang, H. et al. EP300-mediated lysine 2-hydroxyisobutyrylation regulates glycolysis. Mol. Cell 70, 663–678 (2018).
Yang, F., Gao, J., Che, J., Jia, G. & Wang, C. A dimethyl-labeling-based strategy for site-specifically quantitative chemical proteomics. Anal. Chem. 90, 9576–9582 (2018).
Wisniewski, J. R., Zougman, A., Nagaraj, N. & Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 (2009).
Kulevich, S. E., Frey, B. L., Kreitinger, G. & Smith, L. M. Alkylating tryptic peptides to enhance electrospray ionization mass spectrometry analysis. Anal. Chem. 82, 10135–10142 (2010).
Saxon, E. & Bertozzi, C. R. Cell surface engineering by a modified Staudinger reaction. Science 287, 2007–2010 (2000).
Wang, H. et al. Selective in vivo metabolic cell-labeling-mediated cancer targeting. Nat. Chem. Biol. 13, 415–424 (2017).
Acknowledgements
We thank the Computing Platform of the Center for Life Science for supporting the proteomics data analysis and Metabolomics Facility Center in National Protein Science Technology Center of Tsinghua University for C13 isotope tracing experiments. This work is supported by the National Key Research and Development Projects (grant no. 2016YFA0501500) to C.W and X.C., and (no. 2018YFA0507600) to X.C., and the National Natural Science Foundation of China (nos. 21521003, 81490740 and 21778004) to C.W. and (nos. 21425204, 21521003 and 21672013) to X.C., and a ‘1,000 Talents Plan’ Young Investigator Award to C.W.
Author information
Authors and Affiliations
Contributions
W.Q., X.C. and C.W. conceived the project. W.Q. conducted most of the experiments unless specified otherwise. K.Q. carried out the LC–MS/MS analysis. Y.Z. carried out the gene cloning. W.J. cultured BMDMs under the guidance of Y-L.W. Y.C. performed IA-alkyne-based rdTOP-ABPP. B.C. synthesized 1-OH-Az and its derivatives. L.P. helped evaluate the 1-OH-Az labeling. N.C. synthesized UK-alkyne probe. Y.L. analyzed ALDOA structure and W.Z. performed ETD-based LC–MS/MS analysis. W.Q., X.C. and C.W. analyzed the data and wrote the manuscript with input from all the authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–14
Dataset 1
Identification of the cysteines modified by 1-OH-Az and Ac4ManNAz in MCF-7 cells.
Dataset 2
Identification of the proteins modified by 1-OH-Az and Ac4ManNAz in MCF-7 cells.
Dataset 3
Competitive isoTOP-ABPP profiling of itaconate-modified cysteines using the 1-OH-Az probe in Raw264.7 cells with 250 μM of itaconate treatment.
Dataset 4
Competitive isoTOP-ABPP profiling of itaconate-modified cysteines using the 1-OH-Az probe in Raw264.7 cells with 1 mM of itaconate treatment.
Dataset 5
Competitive rdTOP-ABPP profiling of itaconate-modified cysteines using the IA-alkyne probe in Raw264.7 cells.
Rights and permissions
About this article
Cite this article
Qin, W., Qin, K., Zhang, Y. et al. S-glycosylation-based cysteine profiling reveals regulation of glycolysis by itaconate. Nat Chem Biol 15, 983–991 (2019). https://doi.org/10.1038/s41589-019-0323-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-019-0323-5
This article is cited by
-
Neutrophil–macrophage communication via extracellular vesicle transfer promotes itaconate accumulation and ameliorates cytokine storm syndrome
Cellular & Molecular Immunology (2024)
-
Itaconate protects ferroptotic neurons by alkylating GPx4 post stroke
Cell Death & Differentiation (2024)
-
Immunometabolism in atherosclerotic disorders
Nature Cardiovascular Research (2024)
-
Multifaceted mitochondria in innate immunity
npj Metabolic Health and Disease (2024)
-
PMI-controlled mannose metabolism and glycosylation determines tissue tolerance and virus fitness
Nature Communications (2024)