Abstract
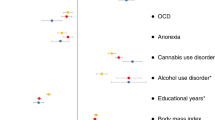
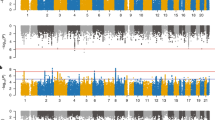
Attention-deficit hyperactivity disorder (ADHD) is a prevalent neurodevelopmental disorder with a major genetic component. Here, we present a genome-wide association study meta-analysis of ADHD comprising 38,691 individuals with ADHD and 186,843 controls. We identified 27 genome-wide significant loci, highlighting 76 potential risk genes enriched among genes expressed particularly in early brain development. Overall, ADHD genetic risk was associated with several brain-specific neuronal subtypes and midbrain dopaminergic neurons. In exome-sequencing data from 17,896 individuals, we identified an increased load of rare protein-truncating variants in ADHD for a set of risk genes enriched with probable causal common variants, potentially implicating SORCS3 in ADHD by both common and rare variants. Bivariate Gaussian mixture modeling estimated that 84–98% of ADHD-influencing variants are shared with other psychiatric disorders. In addition, common-variant ADHD risk was associated with impaired complex cognition such as verbal reasoning and a range of executive functions, including attention.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Summary statistics from the ADHD GWAS meta-analysis are available for download at the PGC website (https://www.med.unc.edu/pgc/download-results/). All relevant iPSYCH data are available from the authors after approval by the iPSYCH Data Access Committee and can only be accessed on the secured Danish server (GenomeDK; https://genome.au.dk) as the data are protected by Danish legislation. For data access and correspondence, please contact D.D. (ditte@biomed.au.dk) or A.D.B. (anders@biomed.au.dk).
Code availability
No previously unreported custom computer code or algorithm was used to generate results.
Change history
01 March 2023
A Correction to this paper has been published: https://doi.org/10.1038/s41588-023-01350-w
References
Faraone, S. V. et al. Attention-deficit/hyperactivity disorder. Nat. Rev. Dis. Prim. 1, 15020 (2015).
Franke, B. et al. The genetics of attention deficit/hyperactivity disorder in adults, a review. Mol. Psychiatry 17, 960–987 (2012).
Dalsgaard, S., Leckman, J. F., Mortensen, P. B., Nielsen, H. S. & Simonsen, M. Effect of drugs on the risk of injuries in children with attention deficit hyperactivity disorder: a prospective cohort study. Lancet Psychiatry 2, 702–709 (2015).
Chang, Z., Lichtenstein, P., D’Onofrio, B. M., Sjolander, A. & Larsson, H. Serious transport accidents in adults with attention-deficit/hyperactivity disorder and the effect of medication: a population-based study. JAMA Psychiatry 71, 319–325 (2014).
Babinski, D. E., Neely, K. A., Ba, D. M. & Liu, G. Depression and suicidal behavior in young adult men and women with ADHD: evidence from claims data. J. Clin. Psychiatry 81, 19m13130 (2020).
Capusan, A. J., Bendtsen, P., Marteinsdottir, I. & Larsson, H. Comorbidity of adult ADHD and its subtypes with substance use disorder in a large population-based epidemiological study. J. Atten. Disord. 23, 1416–1426 (2019).
Boomsma, D. I., van Beijsterveldt, T., Odintsova, V. V., Neale, M. C. & Dolan, C. V. Genetically informed regression analysis: application to aggression prediction by inattention and hyperactivity in children and adults. Behav. Genet. 51, 250–263 (2021).
Dalsgaard, S., Ostergaard, S. D., Leckman, J. F., Mortensen, P. B. & Pedersen, M. G. Mortality in children, adolescents, and adults with attention deficit hyperactivity disorder: a nationwide cohort study. Lancet 385, 2190–2196 (2015).
Jangmo, A. et al. Attention-deficit/hyperactivity disorder and occupational outcomes: the role of educational attainment, comorbid developmental disorders, and intellectual disability. PLoS ONE 16, e0247724 (2021).
Zhao, X. et al. Family burden of raising a child with ADHD. J. Abnorm. Child Psychol. 47, 1327–1338 (2019).
Le, H. H. et al. Economic impact of childhood/adolescent ADHD in a European setting: the Netherlands as a reference case. Eur. Child Adolesc. Psychiatry 23, 587–598 (2014).
Libutzki, B. et al. Direct medical costs of ADHD and its comorbid conditions on basis of a claims data analysis. Eur. Psychiatry 58, 38–44 (2019).
Faraone, S. V. & Larsson, H. Genetics of attention deficit hyperactivity disorder. Mol. Psychiatry 24, 562–575 (2019).
Demontis, D. et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 51, 63–75 (2019).
Pedersen, C. B. et al. The iPSYCH2012 case-cohort sample: new directions for unravelling genetic and environmental architectures of severe mental disorders. Mol. Psychiatry 23, 6–14 (2018).
Mattheisen, M. et al. Identification of shared and differentiating genetic architecture for autism spectrum disorder, attention-deficit hyperactivity disorder and case subgroups. Nat. Genet. 54, 1470–1478 (2022).
Satterstrom, F. K. et al. Autism spectrum disorder and attention deficit hyperactivity disorder have a similar burden of rare protein-truncating variants. Nat. Neurosci. 22, 1961–1965 (2019).
Mullins, N. et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat. Genet. 53, 817–829 (2021).
Howard, D. M. et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci. 22, 343–352 (2019).
Pardinas, A. F. et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet. 50, 381–389 (2018).
Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Trzaskowski, M. et al. Quantifying between-cohort and between-sex genetic heterogeneity in major depressive disorder. Am. J. Med. Genet. B Neuropsychiatr. Genet. 180, 439–447 (2019).
Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017).
Koopmans, F. et al. SynGO: an evidence-based, expert-curated knowledge base for the synapse. Neuron 103, 217–234.e4 (2019).
Xie, Z. et al. Gene set knowledge discovery with enrichr. Curr. Protoc. 1, e90 (2021).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016).
de Leeuw, C. A., Mooij, J. M., Heskes, T. & Posthuma, D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 11, e1004219 (2015).
Zhang, W. et al. Integrative transcriptome imputation reveals tissue-specific and shared biological mechanisms mediating susceptibility to complex traits. Nat. Commun. 10, 3834 (2019).
Wang, D. et al. Comprehensive functional genomic resource and integrative model for the human brain. Science 362, eaat8464 (2018).
Finucane, H. K. et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet. 50, 621–629 (2018).
Meuleman, W. et al. Index and biological spectrum of human DNase I hypersensitive sites. Nature 584, 244–251 (2020).
Corces, M. R. et al. Single-cell epigenomic analyses implicate candidate causal variants at inherited risk loci for Alzheimer’s and Parkinson’s diseases. Nat. Genet. 52, 1158–1168 (2020).
Watanabe, K., Umicevic Mirkov, M., de Leeuw, C. A., van den Heuvel, M. P. & Posthuma, D. Genetic mapping of cell type specificity for complex traits. Nat. Commun. 10, 3222 (2019).
La Manno, G. et al. Molecular diversity of midbrain development in mouse, human, and stem cells. Cell 167, 566–580.e19 (2016).
Zheng, J. et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics 33, 272–279 (2017).
Frei, O. et al. Bivariate causal mixture model quantifies polygenic overlap between complex traits beyond genetic correlation. Nat. Commun. 10, 2417 (2019).
Franke, B. et al. Live fast, die young? A review on the developmental trajectories of ADHD across the lifespan. Eur. Neuropsychopharmacol. 28, 1059–1088 (2018).
Satterthwaite, T. D. et al. Neuroimaging of the Philadelphia Neurodevelopmental Cohort. Neuroimage 86, 544–553 (2014).
Calkins, M. E. et al. The Philadelphia Neurodevelopmental Cohort: constructing a deep phenotyping collaborative. J. Child Psychol. Psychiatry 56, 1356–1369 (2015).
Gur, R. C. et al. Age group and sex differences in performance on a computerized neurocognitive battery in children age 8-21. Neuropsychology 26, 251–265 (2012).
Wilkinson, G. S. & Robertson, G. J. Wide Range Achievement Test (WRAT4) (Psychological Assessment Resources, 2006).
Uffelmann, E. et al. Genome-wide association studies. Nat. Rev. Methods Prim. 1, 59 (2021).
Bataillon, T. et al. The effective size of the Icelandic population and the prospects for LD mapping: inference from unphased microsatellite markers. Eur. J. Hum. Genet. 14, 1044–1053 (2006).
Gazal, S. et al. Linkage disequilibrium-dependent architecture of human complex traits shows action of negative selection. Nat. Genet. 49, 1421–1427 (2017).
Hindley, G. et al. The shared genetic basis of mood instability and psychiatric disorders: a cross-trait genome-wide association analysis. Am. J. Med. Genet. B Neuropsychiatr. Genet. 189, 207–218 (2022).
Plana-Ripoll, O. et al. Exploring comorbidity within mental disorders among a Danish national population. JAMA Psychiatry 76, 259–270 (2019).
Zablotsky, B., Bramlett, M. D. & Blumberg, S. J. The co-occurrence of autism spectrum disorder in children with ADHD. J. Atten. Disord. 24, 94–103 (2020).
Jensen, C. M. & Steinhausen, H. C. Comorbid mental disorders in children and adolescents with attention-deficit/hyperactivity disorder in a large nationwide study. Atten. Defic. Hyperact. Disord. 7, 27–38 (2015).
Chen, Q. et al. Common psychiatric and metabolic comorbidity of adult attention-deficit/hyperactivity disorder: a population-based cross-sectional study. PLoS ONE 13, e0204516 (2018).
Cross-Disorder Group of the Psychiatric Genomics Consortium. Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell 179, 1469–1482.e11 (2019).
Lee, J. J. et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet. 50, 1112–1121 (2018).
Akaike, H. A new look at the statistical model identification. IEEE Trans. Autom. Controls 19, 716–723 (1974).
Yao, X. et al. Integrative analysis of genome-wide association studies identifies novel loci associated with neuropsychiatric disorders. Transl. Psychiatry 11, 69 (2021).
Trubetskoy, V. et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 604, 502–508 (2022).
Johnson, E. C. et al. A large-scale genome-wide association study meta-analysis of cannabis use disorder. Lancet Psychiatry 7, 1032–1045 (2020).
Araujo, D. J. et al. FoxP1 orchestration of ASD-relevant signaling pathways in the striatum. Genes Dev. 29, 2081–2096 (2015).
Fong, W. L., Kuo, H. Y., Wu, H. L., Chen, S. Y. & Liu, F. C. Differential and overlapping pattern of Foxp1 and Foxp2 expression in the striatum of adult mouse brain. Neuroscience 388, 214–223 (2018).
Sollis, E. et al. Equivalent missense variant in the FOXP2 and FOXP1 transcription factors causes distinct neurodevelopmental disorders. Hum. Mutat. 38, 1542–1554 (2017).
Mostafavi, H., Spence, J. P., Naqvi, S. & Pritchard, J. K. Limited overlap of eQTLs and GWAS hits due to systematic differences in discovery. Preprint at bioRxiv https://doi.org/10.1101/2022.05.07.491045 (2022).
Satterstrom, F. K. et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell 180, 568–584.e23 (2020).
Singh, T. et al. Rare coding variants in ten genes confer substantial risk for schizophrenia. Nature 604, 509–516 (2022).
Sazonovs, A. et al. Large-scale sequencing identifies multiple genes and rare variants associated with Crohn’s disease susceptibility. Nat. Genet. 54, 1275–1283 (2021).
Bahmani, Z. et al. Prefrontal contributions to attention and working memory. Curr. Top. Behav. Neurosci. 41, 129–153 (2019).
Sonne, J., Reddy, V. & Beato, M. R. Substantia nigra. in StatPearls (StatPearls Publishing, 2021).
Morales, M. & Margolis, E. B. Ventral tegmental area: cellular heterogeneity, connectivity and behaviour. Nat. Rev. Neurosci. 18, 73–85 (2017).
Chang, S., Yang, L., Wang, Y. & Faraone, S. V. Shared polygenic risk for ADHD, executive dysfunction and other psychiatric disorders. Transl. Psychiatry 10, 182 (2020).
Nigg, J. T. et al. Working memory and vigilance as multivariate endophenotypes related to common genetic risk for attention-deficit/hyperactivity disorder. J. Am. Acad. Child Adolesc. Psychiatry 57, 175–182 (2018).
Aguilar-Lacasana, S. et al. Polygenic risk for ADHD and ASD and their relation with cognitive measures in school children. Psychol. Med. 52, 1356–1364 (2022).
Martin, J., Hamshere, M. L., Stergiakouli, E., O’Donovan, M. C. & Thapar, A. Neurocognitive abilities in the general population and composite genetic risk scores for attention-deficit hyperactivity disorder. J. Child Psychol. Psychiatry 56, 648–656 (2015).
Bybjerg-Grauholm, J. et al. The iPSYCH2015 case-cohort sample: updated directions for unravelling genetic and environmental architectures of severe mental disorders. Preprint at medRxiv https://doi.org/10.1101/2020.11.30.20237768 (2020).
Mors, O., Perto, G. P. & Mortensen, P. B. The Danish psychiatric central research register. Scand. J. Public Health 39, 54–57 (2011).
Lynge, E., Sandegaard, J. L. & Rebolj, M. The Danish national patient register. Scand. J. Public Health 39, 30–33 (2011).
Price, A. L. et al. The impact of divergence time on the nature of population structure: an example from Iceland. PLoS Genet. 5, e1000505 (2009).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Benner, C. et al. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics 32, 1493–1501 (2016).
Greenbaum, J. & Deng, H. W. A statistical approach to fine mapping for the identification of potential causal variants related to bone mineral density. J. Bone Miner. Res. 32, 1651–1658 (2017).
Chen, W. et al. Fine mapping causal variants with an approximate Bayesian method using marginal test statistics. Genetics 200, 719–736 (2015).
Wang, J. et al. CAUSALdb: a database for disease/trait causal variants identified using summary statistics of genome-wide association studies. Nucleic Acids Res. 48, D807–D816 (2020).
Bulik-Sullivan, B. K. et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Gandal, M. J. et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 362, eaat8127 (2018).
Das, S. et al. Next-generation genotype imputation service and methods. Nat. Genet. 48, 1284–1287 (2016).
McCarthy, S. et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 48, 1279–1283 (2016).
Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
Finucane, H. K. et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 47, 1228–1235 (2015).
Liu, M. et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat. Genet. 51, 237–244 (2019).
Grove, J. et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 51, 431–444 (2019).
Yengo, L. et al. Meta-analysis of genome-wide association studies for height and body mass index in approximately 700000 individuals of European ancestry. Hum. Mol. Genet. 27, 3641–3649 (2018).
Okbay, A. et al. Polygenic prediction of educational attainment within and between families from genome-wide association analyses in 3 million individuals. Nat. Genet. 54, 437–449 (2022).
Mills, M. C. et al. Identification of 371 genetic variants for age at first sex and birth linked to externalising behaviour. Nat. Hum. Behav. 5, 1717–1730 (2021).
Watanabe, K. et al. Genome-wide meta-analysis of insomnia prioritizes genes associated with metabolic and psychiatric pathways. Nat. Genet. 54, 1125–1132 (2022).
Als, T. D. et al. Identification of 64 new risk loci for major depression, refinement of the genetic architecture and risk prediction of recurrence and comorbidities. Preprint at medRxiv https://doi.org/10.1101/2022.08.24.22279149 (2022).
Ge, T., Chen, C. Y., Ni, Y., Feng, Y. A. & Smoller, J. W. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun. 10, 1776 (2019).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
Acknowledgements
We thank additional members of the ADHD working group of the PGC who are not named as coauthors under the working group banner for their contributions. We would like to thank the employees and research participants of 23andMe for making this work possible. D.D. was supported by the Novo Nordisk Foundation (NNF20OC0065561), the Lundbeck Foundation (R344-2020-1060) and the European Union’s Horizon 2020 research and innovation program under grant agreement no. 965381 (TIMESPAN). The iPSYCH team was supported by grants from the Lundbeck Foundation (R102-A9118, R155-2014-1724 and R248-2017-2003), National Institutes of Health (NIH)/National Institute of Mental Health (NIMH) (1U01MH109514-01 and 1R01MH124851-01 to A.D.B.) and the Universities and University Hospitals of Aarhus and Copenhagen. The Danish National Biobank resource was supported by the Novo Nordisk Foundation. High-performance computer capacity for handling and statistical analysis of iPSYCH data on the GenomeDK HPC facility was provided by the Center for Genomics and Personalized Medicine and the Centre for Integrative Sequencing, iSEQ, Aarhus University, Denmark (grant to A.D.B.). Research reported in this publication was supported by the NIMH of the NIH under award number R01MH124851. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. B.F. was also supported by funding from the European Community’s Horizon 2020 Programme (H2020/2014 – 2020) under grant agreements no. 728018 (Eat2beNICE) and no. 847879 (PRIME). B.F. also received relevant funding from the Netherlands Organization for Scientific Research for the Dutch National Science Agenda NeurolabNL project (grant 400-17-602). S.E.M. was funded by National Health and Medical Research Council grants APP1172917, APP1158125 and APP1103623. This work was supported by the Instituto de Salud Carlos III (PI19/01224, PI20/0004); by the Pla Estratègic de Recerca i Innovació en Salut, Generalitat de Catalunya (METAL-Cat; SLT006/17/287); by the Agència de Gestió d’Ajuts Universitaris i de Recerca AGAUR, Generalitat de Catalunya (2017SGR1461), Ministry of Science, Innovation and Universities (IJC2018-035346-I to M.S.A.); by the European Regional Development Fund and by ‘la Marató de TV3’ (092330/31) and the European College of Neuropsychopharmacology Network ‘ADHD across the Lifespan’ (https://www.ecnp.eu/researchinnovation/ECNP-networks/List-ECNP-Networks/). T.Z. was funded by NIH, grant no. R37MH107649-07S1 and by Research Council of Norway, NRC, Grant No. 288083. This study was also supported by the NIH, Bethesda, MD, under award numbers T32MH087004 (to K.T.), K08MH122911 (to G.V.), R01MH125246 (to P.R.) and U01MH116442 (to P.R.).
Author information
Authors and Affiliations
Consortia
Contributions
D.D., G.B.W., G.A., R.W., K.T., L.F., G.V., J.B., B.Z., W.Z., J.D., S.H.M. and T.T.N. performed the analysis. D.D., G.B.W., J.G., T.D.A., J.D., F.K.S., J.B.-G., M.B.-H., O.O.G., G.B., K.D., G.S.H., ADHD Working Group of the PGC, iPSYCH-Broad Consortium, E.A., G.E.H., M.N., O.M., D.M.H., P.B.M., M.J.D., H.S., T.W., B.M.N., K.S. and A.D.B. performed sample and/or data provision and processing. D.D., G.B.W., K.T., G.A., G.V., J.B., H.S. and A.D.B. wrote the manuscript. D.D., G.B.W., G.A., R.W., K.T., G.V., J.B., S.D., J.M., M.R., F.K.S., D.I.B., M.S.A., N.R.M., D.H., S.E.M., T.Z., V.M.R., S.V.F., H.S., P.R., B.F., B.M.N., K.S. and A.D.B. performed core revision of the manuscript. D.D. and A.D.B. provided study direction. D.D., G.B.W., H.S., P.R., B.F., T.W., B.M.N., K.S. and A.D.B. supervised the study. All authors contributed to critical revision of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
B.M.N. currently serves as a member of the scientific advisory board at Deep Genomics and Neumora (previously RBNC) and as a consultant for Camp4 Therapeutics, Takeda Pharmaceutical and Biogen. All deCODE-affiliated authors are employees of deCODE/Amgen.
Peer review
Peer review information
Nature Genetics thanks Andrew McQuillin and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Note and Figs. 1–14.
Supplementary Data 1
Regional association plots.
Supplementary Data 2
Forest plots.
Supplementary Tables
Supplementary Tables 1–20.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Demontis, D., Walters, G.B., Athanasiadis, G. et al. Genome-wide analyses of ADHD identify 27 risk loci, refine the genetic architecture and implicate several cognitive domains. Nat Genet 55, 198–208 (2023). https://doi.org/10.1038/s41588-022-01285-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-022-01285-8
This article is cited by
-
Complex interplay of neurodevelopmental disorders (NDDs), fractures, and osteoporosis: a mendelian randomization study
BMC Psychiatry (2024)
-
Identifying causal associations between women’s reproductive traits and risk of schizophrenia: a multivariate validated two-sample Mendelian randomization analysis
BMC Psychiatry (2024)
-
Persistent thinness and anorexia nervosa differ on a genomic level
European Journal of Human Genetics (2024)
-
Cortico-striatal differences in the epigenome in attention-deficit/ hyperactivity disorder
Translational Psychiatry (2024)
-
Causal influences of neuropsychiatric disorders on Alzheimer’s disease
Translational Psychiatry (2024)