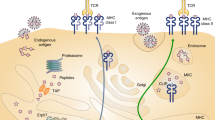
Abstract
The identification of actionable tumor antigens is indispensable for the development of several cancer immunotherapies, including T cell receptor–transduced T cells and patient-specific mRNA or peptide vaccines. Most known tumor antigens have been identified through extensive molecular characterization and are considered canonical if they derive from protein-coding regions of the genome. By eluting human leukocyte antigen-bound peptides from tumors and subjecting these to mass spectrometry analysis, the peptides can be identified by matching the resulting spectra against reference databases. Recently, mass-spectrometry-based immunopeptidomics has enabled the discovery of noncanonical antigens—antigens derived from sequences outside protein-coding regions or generated by noncanonical antigen-processing mechanisms. Coupled with transcriptomics and ribosome profiling, this method enables the identification of thousands of noncanonical peptides, of which a substantial fraction may be detected exclusively in tumors. Spectral matching against the immense noncanonical reference may generate false positives. However, sensitive mass spectrometry, analytical validation and advanced bioinformatics solutions are expected to uncover the full landscape of presented antigens and clinically relevant targets.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Kloetzel, P. M. Antigen processing by the proteasome. Nat. Rev. Mol. Cell Biol. 2, 179–187 (2001).
Coulie, P. G. et al. A mutated intron sequence codes for an antigenic peptide recognized by cytolytic T lymphocytes on a human melanoma. Proc. Natl Acad. Sci. USA 92, 7976–7980 (1995).
Roche, P. A. & Furuta, K. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat. Rev. Immunol. 15, 203–216 (2015).
Yewdell, J. W., Reits, E. & Neefjes, J. Making sense of mass destruction: quantitating MHC class I antigen presentation. Nat. Rev. Immunol. 3, 952–961 (2003).
Schumacher, T. N., Scheper, W. & Kvistborg, P. Cancer neoantigens. Annu. Rev. Immunol. 37, 173–200 (2019).
Bianchi, V., Harari, A. & Coukos, G. Neoantigen-specific adoptive cell therapies for cancer: making T-cell products more personal. Front. Immunol. 11, 1215 (2020).
Curran, M. A. & Glisson, B. S. New hope for therapeutic cancer vaccines in the era of immune checkpoint modulation. Annu. Rev. Med. 70, 409–424 (2019).
Haen, S. P., Löffler, M. W., Rammensee, H.-G. & Brossart, P. Towards new horizons: characterization, classification and implications of the tumour antigenic repertoire. Nat. Rev. Clin. Oncol. 17, 595–610 (2020).
Kruger, S. et al. Advances in cancer immunotherapy 2019: latest trends. J. Exp. Clin. Cancer Res. 38, 268 (2019).
Christofi, T., Baritaki, S., Falzone, L., Libra, M. & Zaravinos, A. Current perspectives in cancer immunotherapy. Cancers (Basel) 11, 1472 (2019).
Laumont, C. M. et al. Global proteogenomic analysis of human MHC class I-associated peptides derived from non-canonical reading frames. Nat. Commun. 7, 10238 (2016).
Sebestyen, E. et al. Large-scale analysis of genome and transcriptome alterations in multiple tumors unveils novel cancer-relevant splicing networks. Genome Res. 26, 732–744 (2016).
Zhao, Q. et al. Proteogenomics uncovers a vast repertoire of shared tumor-specific antigens in ovarian cancer. Cancer Immunol. Res. 8, 544–555 (2020).
Ouspenskaia, T. et al. Thousands of novel unannotated proteins expand the MHC I immunopeptidome in cancer. Preprint at bioRxiv https://doi.org/10.1101/2020.02.12.945840 (2020).
Chen, J. et al. Pervasive functional translation of noncanonical human open reading frames. Science 367, 1140–1146 (2020).
Ilyas, S. & Yang, J. C. Landscape of tumor antigens in T cell immunotherapy. J. Immunol. 195, 5117–5122 (2015).
Caballero, O. L. & Chen, Y. T. Cancer/testis (CT) antigens: potential targets for immunotherapy. Cancer Sci. 100, 2014–2021 (2009).
Tio, D. et al. Expression of cancer/testis antigens in cutaneous melanoma: a systematic review. Melanoma Res. 29, 349–357 (2019).
Schooten, E., Di Maggio, A., van Bergen En Henegouwen, P. M. P. & Kijanka, M. M. MAGE-A antigens as targets for cancer immunotherapy. Cancer Treat. Rev. 67, 54–62 (2018).
D’Angelo, S. P. et al. Antitumor activity associated with prolonged persistence of adoptively transferred NY-ESO-1 c259T cells in synovial. Sarcoma 8, 944–957 (2018).
Rapoport, A. P. et al. NY-ESO-1–specific TCR–engineered T cells mediate sustained antigen-specific antitumor effects in myeloma. Nat. Med. 21, 914–921 (2015).
Robbins, P. F. et al. A pilot trial using lymphocytes genetically engineered with an NY-ESO-1–reactive T-cell receptor: long-term follow-up and correlates with response. Clin. Cancer Res. 21, 1019–1027 (2015).
Laumont, C. M. & Perreault, C. Exploiting non-canonical translation to identify new targets for T cell-based cancer immunotherapy. Cell. Mol. Life Sci. 75, 607–621 (2018).
Moreau-Aubry, A. et al. A processed pseudogene codes for a new antigen recognized by a CD8+ T cell clone on melanoma. J. Exp. Med. 191, 1617–1623 (2000).
Li, L.-J., Leng, R.-X., Fan, Y.-G., Pan, H.-F. & Ye, D.-Q. Translation of noncoding RNAs: focus on lncRNAs, pri-miRNAs, and circRNAs. Exp. Cell Res. 361, 1–8 (2017).
Charpentier, M. et al. IRES-dependent translation of the long non coding RNA meloe in melanoma cells produces the most immunogenic MELOE antigens. Oncotarget 7, 59704–59713 (2016).
Roulois, D. et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 162, 961–973 (2015).
Chiappinelli, K. B. et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162, 974–986 (2015).
Attermann, A. S., Bjerregaard, A. M., Saini, S. K., Gronbaek, K. & Hadrup, S. R. Human endogenous retroviruses and their implication for immunotherapeutics of cancer. Ann. Oncol. 29, 2183–2191 (2018).
Vigneron, N. et al. An antigenic peptide produced by peptide splicing in the proteasome. Science 304, 587–590 (2004).
Delong, T. et al. Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science 351, 711–714 (2016).
Yewdell, J. W. & Holly, J. DRiPs get molecular. Curr. Opin. Immunol. 64, 130–136 (2020).
Welters, M. J. et al. Induction of tumor-specific CD4+ and CD8+ T-cell immunity in cervical cancer patients by a human papillomavirus type 16 E6 and E7 long peptides vaccine. Clin. Cancer Res. 14, 178–187 (2008).
Morgan, R. A. et al. Cancer regression and neurological toxicity following anti-MAGE-A3 TCR gene therapy. J. Immunother. 36, 133–151 (2013).
Skipper, J. C. et al. Mass-spectrometric evaluation of HLA-A*0201-associated peptides identifies dominant naturally processed forms of CTL epitopes from MART-1 and gp100. Int J. Cancer 82, 669–677 (1999).
Wolf, B. et al. Safety and tolerability of adoptive cell therapy in cancer. Drug Saf. 42, 315–334 (2019).
Purcell, A. W., Ramarathinam, S. H. & Ternette, N. Mass spectrometry-based identification of MHC-bound peptides for immunopeptidomics. Nat. Protoc. 14, 1687–1707 (2019).
Caron, E. et al. Analysis of major histocompatibility complex (MHC) immunopeptidomes using mass spectrometry. Mol. Cell. Proteom. 14, 3105–3117 (2015).
Ritz, D., Kinzi, J., Neri, D. & Fugmann, T. Data-independent acquisition of HLA class I peptidomes on the Q exactive mass spectrometer platform. Proteomics 17, 1700177 (2017).
Gillet, L. C. et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteom. 11, O111.016717 (2012).
Brunner, A.-D. et al. Ultra-high sensitivity mass spectrometry quantifies single-cell proteome changes upon perturbation. Preprint at bioRxiv 2020.2012.2022.423933 (2020).
Tsou, C. C. et al. DIA-Umpire: comprehensive computational framework for data-independent acquisition proteomics. Nat. Methods 12, 258–264 (2015).
Muntel, J. et al. Surpassing 10 000 identified and quantified proteins in a single run by optimizing current LC-MS instrumentation and data analysis strategy. Mol. Omics 15, 348–360 (2019).
Gessulat, S. et al. Prosit: proteome-wide prediction of peptide tandem mass spectra by deep learning. Nat. Methods 16, 509–518 (2019).
Croft, N. P. et al. Kinetics of antigen expression and epitope presentation during virus infection. PLoS Pathog. 9, e1003129 (2013).
Hassan, C. et al. Accurate quantitation of MHC-bound peptides by application of isotopically labeled peptide MHC complexes. J. Proteom. 109, 240–244 (2014).
Croft, N. P., Purcell, A. W. & Tscharke, D. C. Quantifying epitope presentation using mass spectrometry. Mol. Immunol. 68, 77–80 (2015).
Tan, C. T., Croft, N. P., Dudek, N. L., Williamson, N. A. & Purcell, A. W. Direct quantitation of MHC-bound peptide epitopes by selected reaction monitoring. Proteomics 11, 2336–2340 (2011).
Kapp, E. A. et al. An evaluation, comparison, and accurate benchmarking of several publicly available MS/MS search algorithms: sensitivity and specificity analysis. Proteomics 5, 3475–3490 (2005).
Kapp, E. & Schutz, F. Overview of tandem mass spectrometry (MS/MS) database search algorithms. Curr. Protoc. Protein Sci. 49, 25.2.1–25.2.19 (2007).
Elias, J. E. & Gygi, S. P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 (2007).
Zhang, J. et al. PEAKS DB: de novo sequencing assisted database search for sensitive and accurate peptide identification. Mol. Cell. Proteom. 11, M111.010587 (2012).
Shan, P. & Tran, H. Integrating database search and de novo sequencing for immunopeptidomics with DIA approach. J. Biomol. Tech. 30, S23 (2019).
Faridi, P., Purcell, A. W. & Croft, N. P. In immunopeptidomics we need a sniper instead of a shotgun. Proteomics 18, e1700464 (2018).
Thompson, A. et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 75, 1895–1904 (2003).
Pfammatter, S. et al. Extending the comprehensiveness of immunopeptidome analyses using isobaric peptide labeling. Anal. Chem. 92, 9194–9204 (2020).
Ramarathinam, S. H. et al. A peptide-signal amplification strategy for the detection and validation of neoepitope presentation on cancer biopsies. Preprint at bioRxiv https://doi.org/10.1101/2020.06.12.145276 (2020).
Stopfer, L. E., Mesfin, J. M., Joughin, B. A., Lauffenburger, D. A. & White, F. M. Multiplexed relative and absolute quantitative immunopeptidomics reveals MHC I repertoire alterations induced by CDK4/6 inhibition. Nat. Commun. 11, 2760 (2020).
d’Atri, V. et al. Adding a new separation dimension to MS and LC–MS: what is the utility of ion mobility spectrometry? J. Sep. Sci. 41, 20–67 (2018).
Pfammatter, S. et al. A novel differential ion mobility device expands the depth of proteome coverage and the sensitivity of multiplex proteomic measurements. Mol. Cell. Proteom. 17, 2051–2067 (2018).
Pfammatter, S., Bonneil, E. & Thibault, P. Improvement of quantitative measurements in multiplex proteomics using high-field asymmetric waveform spectrometry. J. Proteome Res. 15, 4653–4665 (2016).
Meier, F. et al. Online parallel accumulation-serial fragmentation (PASEF) with a novel trapped ion mobility mass spectrometer. Mol. Cell. Proteom. 17, 2534–2545 (2018).
Nesvizhskii, A. I. Proteogenomics: concepts, applications and computational strategies. Nat. Methods 11, 1114–1125 (2014).
Zhang, M. et al. RNA editing derived epitopes function as cancer antigens to elicit immune responses. Nat. Commun. 9, 3919 (2018).
Wei, Z. et al. The landscape of tumor fusion neoantigens: a pan-cancer. Anal. iScience 21, 249–260 (2019).
Löffler, M. W. et al. Multi-omics discovery of exome-derived neoantigens in hepatocellular carcinoma. Genome Med. 11, 28 (2019).
Kalaora, S. et al. Use of HLA peptidomics and whole exome sequencing to identify human immunogenic neo-antigens. Oncotarget 7, 5110–5117 (2016).
Bassani-Sternberg, M., Pletscher-Frankild, S., Jensen, L. J. & Mann, M. Mass spectrometry of human leukocyte antigen class I peptidomes reveals strong effects of protein abundance and turnover on antigen presentation. Mol. Cell. Proteom. 14, 658–673 (2015).
Khodadoust, M. S. et al. Antigen presentation profiling reveals recognition of lymphoma immunoglobulin neoantigens. Nature 543, 723–727 (2017).
Bassani-Sternberg, M. et al. Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat. Commun. 7, 13404 (2016).
Binz, P. A. et al. Proteomics Standards Initiative extended FASTA format. J. Proteome Res. 18, 2686–2692 (2019).
Eng, J. K. & Deutsch, E. W. Extending comet for global amino acid variant and post-translational modification analysis using the PSI extended FASTA format. Proteomics 72, e1900362 (2020).
Elias, J. E. & Gygi, S. P. Target-decoy search strategy for mass spectrometry-based proteomics. Methods Mol. Biol. 604, 55–71 (2010).
Gupta, N., Bandeira, N., Keich, U. & Pevzner, P. A. Target-decoy approach and false discovery rate: when things may go wrong. J. Am. Soc. Mass Spectrom. 22, 1111–1120 (2011).
Tanner, S. et al. Improving gene annotation using peptide mass spectrometry. Genome Res. 17, 231–239 (2007).
Chong, C. et al. Integrated proteogenomic deep sequencing and analytics accurately identify non-canonical peptides in tumor immunopeptidomes. Nat. Commun. 11, 1293 (2020).
Laumont, C. M. et al. Noncoding regions are the main source of targetable tumor-specific antigens. Sci. Transl. Med. 10, eaau5516 (2018).
Smart, A. C. et al. Intron retention is a source of neoepitopes in cancer. Nat. Biotechnol. 36, 1056–1058 (2018).
Attig, J. et al. LTR retroelement expansion of the human cancer transcriptome and immunopeptidome revealed by de novo transcript assembly. Genome Res. 29, 1578–1590 (2019).
Kong, Y. et al. Transposable element expression in tumors is associated with immune infiltration and increased antigenicity. Nat. Commun. 10, 5228 (2019).
Shraibman, B., Melamed Kadosh, D., Barnea, E. & Admon, A. HLA peptides derived from tumor antigens induced by inhibition of DNA methylation for development of drug-facilitated immunotherapy. Mol. Cell. Proteom. 15, 3058–3070 (2016).
Calviello, L. et al. Detecting actively translated open reading frames in ribosome profiling data. Nat. Methods 13, 165–170 (2016).
Erhard, F. et al. Improved Ribo-seq enables identification of cryptic translation events. Nat. Methods 15, 363–366 (2018).
Ingolia, N. T. et al. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 8, 1365–1379 (2014).
Slavoff, S. A. et al. Peptidomic discovery of short open reading frame–encoded peptides in human cells. Nat. Chem. Biol. 9, 59–64 (2013).
Sarkizova, S. et al. A large peptidome dataset improves HLA class I epitope prediction across most of the human population. Nat. Biotechnol. 38, 199–209 (2020).
Abelin, J. G. et al. Mass spectrometry profiling of HLA-associated peptidomes in mono-allelic cells enables more accurate epitope prediction. Immunity 46, 315–326 (2017).
Warren, E. H. et al. An antigen produced by splicing of noncontiguous peptides in the reverse order. Science 313, 1444–1447 (2006).
Dalet, A. et al. An antigenic peptide produced by reverse splicing and double asparagine deamidation. Proc. Natl Acad. Sci. USA 108, E323–E331 (2011).
Michaux, A. et al. A spliced antigenic peptide comprising a single spliced amino acid is produced in the proteasome by reverse splicing of a longer peptide fragment followed by trimming. J. Immunol. 192, 1962–1971 (2014).
Hanada, K., Yewdell, J. W. & Yang, J. C. Immune recognition of a human renal cancer antigen through post-translational protein splicing. Nature 427, 252–256 (2004).
Liepe, J. et al. A large fraction of HLA class I ligands are proteasome-generated spliced peptides. Science 354, 354–358 (2016).
Faridi, P. et al. A subset of HLA-I peptides are not genomically templated: evidence for cis- and trans-spliced peptide ligands. Sci. Immunol. 3, eaar3947 (2018).
Liepe, J., Sidney, J., Lorenz, F. K. M., Sette, A. & Mishto, M. Mapping the MHC class I–spliced immunopeptidome of cancer cells. Cancer Immunol. Res. 7, 62–76 (2019).
Paes, W. et al. Contribution of proteasome-catalyzed peptide cis-splicing to viral targeting by CD8+ T cells in HIV-1 infection. Proc. Natl Acad. Sci. USA 116, 24748–24759 (2019).
Faridi, P. et al. Spliced peptides and cytokine-driven changes in the immunopeptidome of melanoma. Cancer Immunol. Res. 8, 1322–1334 (2020).
Mylonas, R. et al. Estimating the contribution of proteasomal spliced peptides to the HLA-I ligandome. Mol. Cell. Proteom. 17, 2347–2357 (2018).
Rolfs, Z., Solntsev, S. K., Shortreed, M. R., Frey, B. L. & Smith, L. M. Global identification of post-translationally spliced peptides with neo-fusion. J. Proteome Res. 18, 349–358 (2019).
Erhard, F., Dölken, L., Schilling, B. & Schlosser, A. Identification of the cryptic HLA-I immunopeptidome. Cancer Immunol. Res. 8, 1018–1026 (2020).
Vigneron, N., Ferrari, V., Stroobant, V., Abi Habib, J. & Van den Eynde, B. J. Peptide splicing by the proteasome. J. Biol. Chem. 292, 21170–21179 (2017).
Dalet, A., Vigneron, N., Stroobant, V., Hanada, K. & Van den Eynde, B. J. Splicing of distant peptide fragments occurs in the proteasome by transpeptidation and produces the spliced antigenic peptide derived from fibroblast growth factor-5. J. Immunol. 184, 3016–3024 (2010).
Henry, V. J., Bandrowski, A. E., Pepin, A. S., Gonzalez, B. J. & Desfeux, A. OMICtools: an informative directory for multi-omic data analysis. Database (Oxford) 2014, bau069 (2014).
Afgan, E. et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537–W544 (2018).
Nesvizhskii, A. I. et al. Dynamic spectrum quality assessment and iterative computational analysis of shotgun proteomic data: toward more efficient identification of post-translational modifications, sequence polymorphisms, and novel peptides. Mol. Cell. Proteom. 5, 652–670 (2006).
Andreatta, M. et al. MS-Rescue: a computational pipeline to increase the quality and yield of immunopeptidomics experiments. Proteomics 19, e1800357 (2019).
Rolfs, Z., Müller, M., Shortreed, M. R., Smith, L. M. & Bassani-Sternberg, M. Comment on ‘A subset of HLA-I peptides are not genomically templated: evidence for cis- and trans-spliced peptide ligands’. Sci. Immunol. 4, eaaw1622 (2019).
McGranahan, N. & Swanton, C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 168, 613–628 (2017).
Marcu, A. et al. The HLA Ligand Atlas. A resource of natural HLA ligands presented on benign tissues. J. Immunother. Cancer 9, e002071 (2019).
Schatton, T. et al. Identification of cells initiating human melanomas. Nature 451, 345–349 (2008).
Lang, D., Mascarenhas, J. B. & Shea, C. R. Melanocytes, melanocyte stem cells, and melanoma stem cells. Clin. Dermatol. 31, 166–178 (2013).
Kassiotis, G. & Stoye, J. P. Immune responses to endogenous retroelements: taking the bad with the good. Nat. Rev. Immunol. 16, 207–219 (2016).
Rycaj, K. et al. Cytotoxicity of human endogenous retrovirus K–specific T cells toward autologous ovarian. Cancer Cells 21, 471–483 (2015).
Saini, S. K. et al. Human endogenous retroviruses form a reservoir of T cell targets in hematological cancers. Nat. Commun. 11, 5660 (2020).
Mullins, C. S. & Linnebacher, M. Endogenous retrovirus sequences as a novel class of tumor-specific antigens: an example of HERV-H env encoding strong CTL epitopes. Cancer Immunol. Immun. 61, 1093–1100 (2012).
Tu, X. et al. Human leukemia antigen-A*0201-restricted epitopes of human endogenous retrovirus W family envelope (HERV-W env) induce strong cytotoxic T lymphocyte responses. Virol. Sin. 32, 280–289 (2017).
Belgnaoui, S. M., Gosden, R. G., Semmes, O. J. & Haoudi, A. Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells. Cancer Cell Int. 6, 13 (2006).
Scott, E. C. et al. A hot L1 retrotransposon evades somatic repression and initiates human colorectal cancer. Genome Res. 26, 745–755 (2016).
Ott, P. A. et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547, 217–221 (2017).
Sahin, U. et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 547, 222–226 (2017).
Ebrahimi-Nik, H. et al. Mass spectrometry driven exploration reveals nuances of neoepitope-driven tumor rejection. JCI Insight 5, e129152 (2019).
Smith, C. C. et al. Alternative tumour-specific antigens. Nat. Rev. Cancer 19, 465–478 (2019).
Jackson, R. et al. The translation of non-canonical open reading frames controls mucosal immunity. Nature 564, 434–438 (2018).
Muller, M., Gfeller, D., Coukos, G. & Bassani-Sternberg, M. ‘Hotspots’ of antigen presentation revealed by human leukocyte antigen ligandomics for neoantigen prioritization. Front. Immunol. 8, 1367 (2017).
Schittenhelm, R. B. et al. A comprehensive analysis of constitutive naturally processed and presented HLA-C*04:01 (Cw4)-specific peptides. Tissue Antigen. 83, 174–179 (2014).
Schuster, H. et al. The immunopeptidomic landscape of ovarian carcinomas. Proc. Natl Acad. Sci. USA 114, E9942–E9951 (2017).
Sarkizova, S. et al. A large peptidome dataset improves HLA class I epitope prediction across most of the human population. Nat. Biotechnol. 38, 199–209 (2020).
Keskin, D. B. et al. Neoantigen vaccine generates intratumoral T cell responses in Phase Ib glioblastoma trial. Nature 565, 234–239 (2019).
Shraibman, B. et al. Identification of tumor antigens among the HLA peptidomes of glioblastoma tumors and plasma. Mol. Cell. Proteom. 17, 2132–2145 (2018).
Ternette, N. et al. Immunopeptidomic profiling of HLA-A2-positive triple negative breast cancer identifies potential immunotherapy target antigens. Proteomics 18, 1700465 (2018).
Acknowledgements
This study was supported by the Ludwig Institute for Cancer Research and grant KFS-4680-02-2019-R from the Swiss Cancer League (M.B.-S.). This work was also supported by grants from Cancera, Mats Paulssons and by a gift from the Biltema Foundation that was administered by the ISREC Foundation, Lausanne, Switzerland. The figures were originally created with BioRender.com.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chong, C., Coukos, G. & Bassani-Sternberg, M. Identification of tumor antigens with immunopeptidomics. Nat Biotechnol 40, 175–188 (2022). https://doi.org/10.1038/s41587-021-01038-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-021-01038-8
This article is cited by
-
Immunopeptidomics-based identification of naturally presented non-canonical circRNA-derived peptides
Nature Communications (2024)
-
Tumour circular RNAs elicit anti-tumour immunity by encoding cryptic peptides
Nature (2024)
-
Thunder-DDA-PASEF enables high-coverage immunopeptidomics and is boosted by MS2Rescore with MS2PIP timsTOF fragmentation prediction model
Nature Communications (2024)
-
Overexpression of Transmembrane Phosphatase with Tensin homology (TPTE) in prostate cancer is clinically significant, suggesting its potential as a valuable biomarker
Journal of Cancer Research and Clinical Oncology (2024)
-
ABPEPserver: a web application for documentation and analysis of substitutants
BMC Cancer (2023)