Abstract
Nine dogs with hemophilia A were treated with adeno-associated viral (AAV) gene therapy and followed for up to 10 years. Administration of AAV8 or AAV9 vectors expressing canine factor VIII (AAV-cFVIII) corrected the FVIII deficiency to 1.9–11.3% of normal FVIII levels. In two of nine dogs, levels of FVIII activity increased gradually starting about 4 years after treatment. None of the dogs showed evidence of tumors or altered liver function. Analysis of integration sites in liver samples from six treated dogs identified 1,741 unique AAV integration events in genomic DNA and expanded cell clones in five dogs, with 44% of the integrations near genes involved in cell growth. All recovered integrated vectors were partially deleted and/or rearranged. Our data suggest that the increase in FVIII protein expression in two dogs may have been due to clonal expansion of cells harboring integrated vectors. These results support the clinical development of liver-directed AAV gene therapy for hemophilia A, while emphasizing the importance of long-term monitoring for potential genotoxicity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The raw sequencing data supporting this study are available at the Zenodo data server (https://doi.org/10.5281/zenodo.3666122), while demultiplexed sample reads generated during the analysis are available at the NIH SRA (BioProject ID: PRJNA606282). Source data are provided with this paper.
Code availability
The AAVenger software and analysis software supporting this study are available at the Zenodo data server (https://doi.org/10.5281/zenodo.3666122). Source data are provided with this paper.
References
Rangarajan, S. et al. AAV5-factor VIII gene transfer in severe hemophilia A. N. Engl. J. Med. 377, 2519–2530 (2017).
High, K. A. et al. A phase 1/2 trial of investigational Spk-8011 in hemophilia A demonstrates durable expression and prevention of bleeds. Blood 132, 487 (2018).
Nathwani, A. C. et al. GO-8: preliminary results of a phase I/II dose escalation trial of gene therapy for haemophilia A using a novel human factor VIII variant. Blood 132, 489 (2018).
Pasi, K. J. et al. Multiyear follow-up of AAV5-hFVIII-SQ gene therapy for hemophilia A. N. Engl. J. Med. 382, 29–40 (2020).
Jiang, H. et al. Multiyear therapeutic benefit of AAV serotypes 2, 6, and 8 delivering factor VIII to hemophilia A mice and dogs. Blood 108, 107–115 (2006).
Sabatino, D. E. et al. Efficacy and safety of long-term prophylaxis in severe hemophilia A dogs following liver gene therapy using AAV vectors. Mol. Ther. 19, 442–449 (2011).
Sarkar, R. et al. Long-term efficacy of adeno-associated virus serotypes 8 and 9 in hemophilia A dogs and mice. Hum. Gene Ther. 17, 427–439 (2006).
Nathwani, A. C. et al. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N. Engl. J. Med. 371, 1994–2004 (2014).
Nathwani, A. C. et al. Adeno-associated mediated gene transfer for hemophilia B: 8 year follow up and impact of removing ‘empty viral particles’ on safety and efficacy gene transfer. Blood 132, 491 (2018).
Nakai, H. et al. Extrachromosomal recombinant adeno-associated virus vector genomes are primarily responsible for stable liver transduction in vivo. J. Virol. 75, 6969–6976 (2001).
Schotanus, B. A., Penning, L. C. & Spee, B. Potential of regenerative medicine techniques in canine hepatology. Vet. Q. 33, 207–216 (2013).
Li, H. et al. Assessing the potential for AAV vector genotoxicity in a murine model. Blood 117, 3311–3319 (2011).
Nakai, H. et al. Large-scale molecular characterization of adeno-associated virus vector integration in mouse liver. J. Virol. 79, 3606–3614 (2005).
Chandler, R. J. et al. Vector design influences hepatic genotoxicity after adeno-associated virus gene therapy. J. Clin. Invest. 125, 870–880 (2015).
Chandler, R. J., Sands, M. S. & Venditti, C. P. Recombinant adeno-associated viral integration and genotoxicity: insights from animal models. Hum. Gene Ther. 28, 314–322 (2017).
Zhong, L. et al. Recombinant adeno-associated virus integration sites in murine liver after ornithine transcarbamylase gene correction. Hum. Gene Ther. 24, 520–525 (2013).
Gil-Farina, I. et al. Recombinant AAV integration is not associated with hepatic genotoxicity in nonhuman primates and patients. Mol. Ther. 24, 1100–1105 (2016).
Kaeppel, C. et al. A largely random AAV integration profile after LPLD gene therapy. Nat. Med. 19, 889–891 (2013).
Nault, J.-C. et al. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nat. Genet. 47, 1187–1193 (2015).
La Bella, T. et al. Adeno-associated virus in the liver: natural history and consequences in tumour development. Gut 69, 737–747 (2020).
Büning, H. & Schmidt, M. Adeno-associated vector toxicity—to be or not to be? Mol. Ther. 23, 1673–1675 (2015).
Logan, G. J. et al. Identification of liver-specific enhancer-promoter activity in the 3′ untranslated region of the wild-type AAV2 genome. Nat. Genet. 49, 1267–1273 (2017).
Bell, P. et al. No evidence for tumorigenesis of AAV vectors in a large-scale study in mice. Mol. Ther. 12, 299–306 (2005).
Donsante, A. et al. Observed incidence of tumorigenesis in long-term rodent studies of rAAV vectors. Gene Ther. 8, 1343–1346 (2001).
Donsante, A. et al. AAV vector integration sites in mouse hepatocellular carcinoma. Science 317, 477 (2007).
Walia, J. S. et al. Long-term correction of Sandhoff disease following intravenous delivery of rAAV9 to mouse neonates. Mol. Ther. 23, 414–422 (2016).
Rosas, L. E. et al. Patterns of scAAV vector insertion associated with oncogenic events in a mouse model for genotoxicity. Mol. Ther. 20, 2098–2110 (2012).
Bell, P. et al. Analysis of tumors arising in male B6C3F1 mice with and without AAV vector delivery to liver. Mol. Ther. 14, 34–44 (2006).
Lozier, J. N. et al. The Chapel hill hemophilia A dog colony exhibits a factor VIII gene inversion. Proc. Natl Acad. Sci. USA 99, 12991–12996 (2002).
Sabatino, D. E. et al. Recombinant canine B-domain-deleted FVIII exhibits high specific activity and is safe in the canine hemophilia A model. Blood 114, 4562–4565 (2009).
McCormack, W. M. et al. Helper-dependent adenoviral gene therapy mediates long-term correction of the clotting defect in the canine hemophilia A model. J. Thromb. Haemost. 4, 1218–1225 (2006).
Berntorp, E., Spotts, G., Patrone, L. & Ewenstein, B. M. Advancing personalized care in hemophilia A: ten years’ experience with an advanced category antihemophilic factor prepared using a plasma/albumin-free method. Biologics 8, 115–127 (2014).
Center, S. A. Interpretation of liver enzymes. Vet. Clin. North Am. Small Anim. Pract. 37, 297–333 (2007).
Galle, P. R. et al. Biology and significance of α-fetoprotein in hepatocellular carcinoma. Liver Int. 39, 2214–2229 (2019).
Kitao, S. et al. α-fetoprotein in serum and tumor tissues in dogs with hepatocellular carcinoma. J. Vet. Diagn. Invest. 18, 291–295 (2006).
Yamada, T. et al. Serum α-fetoprotein values in dogs with various hepatic diseases. J. Vet. Med. Sci. 61, 657–659 (1999).
Berry, C. C. et al. Estimating abundances of retroviral insertion sites from DNA fragment length data. Bioinformatics 28, 755–762 (2012).
Yang, C. C. et al. Cellular recombination pathways and viral terminal repeat hairpin structures are sufficient for adeno-associated virus integration in vivo and in vitro. J. Virol. 71, 9231–9247 (1997).
Gaidano, G., Foà, R. & Dalla-Favera, R. Molecular pathogenesis of chronic lymphocytic leukemia. J. Clin. Invest. 122, 3432–3438 (2012).
Huang, R. Q. et al. Knockdown of PEBP4 inhibits human glioma cell growth and invasive potential via ERK1/2 signaling pathway. Mol. Carcinog. 58, 135–143 (2019).
Zhang, D. et al. PEBP4 promoted the growth and migration of cancer cells in pancreatic ductal adenocarcinoma. Tumour Biol. 37, 1699–1705 (2016).
Berry, C. C., Ocwieja, K. E., Malani, N. & Bushman, F. D. Comparing DNA integration site clusters with scan statistics. Bioinformatics 30, 1493–1500 (2014).
Cogné, B. et al. NGS library preparation may generate artifactual integration sites of AAV vectors. Nat. Med. 20, 577–578 (2014).
Kao, C.-Y. et al. Incorporation of the factor IX Padua mutation into FIX-Triple improves clotting activity in vitro and in vivo. Thromb. Haemost. 110, 244–256 (2013).
Samulski, R. J., Chang, L. S. & Shenk, T. A recombinant plasmid from which an infectious adeno-associated virus genome can be excised in vitro and its use to study viral replication. J. Virol. 61, 3096–3101 (1987).
Donne, R., Saroul-Aïnama, M., Cordier, P., Celton-Morizur, S. & Desdouets, C. Polyploidy in liver development, homeostasis and disease. Nat. Rev. Gastroenterol. Hepatol. 17, 391–405 (2020).
Kyrle, P. A. et al. High plasma levels of factor VIII and the risk of recurrent venous thromboembolism. N. Engl. J. Med. 343, 457–462 (2000).
Rietveld, I. M. et al. High levels of coagulation factors and venous thrombosis risk: strongest association for factor VIII and von Willebrand factor. J. Thromb. Haemost. 17, 99–109 (2019).
Sadelain, M., Papapetrou, E. P. & Bushman, F. D. Safe harbours for the integration of new DNA in the human genome. Nat. Rev. Cancer 12, 51–58 (2011).
Sherman, A. et al. Portal vein delviery of viral vectors for gene therapy for hemophilia. Methods Mol. Biol. 1114, 413–426 (2014).
Hothorn, T., Hornik, K., van de Wiel, M. & Zeileis, A. Implementing a class of permutation tests: the coin package. J. Stat. Softw. 28, 1–23 (2008).
Sherman, E. et al. INSPIIRED: a pipeline for quantitative analysis of sites of new DNA integration in cellular genomes. Mol. Ther. Methods Clin. Dev. 4, 39–49 (2017).
Berry, C. C. et al. INSPIIRED: quantification and visualization tools for analyzing integration site distributions. Mol. Ther. Methods Clin. Dev. 4, 17–26 (2017).
Pruitt, K. D., Tatusova, T. & Maglott, D. R. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 33, D501–D504 (2005).
Pruitt, K. D. et al. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 42, D756–D763 (2014).
Kent, W. J. BLAT—the BLAST-like alignment tool. Genome Res. 12, 656–664 (2002).
Acknowledgements
We are grateful to members of the Sabatino and Bushman laboratories for help and suggestions. We acknowledge the Research Vector Core at the Children’s Hospital of Philadelphia for production of the SC AAV vectors and the Penn Vector Core at the University of Pennsylvania for preparing the TC AAV vectors. We thank M. Keiser for assistance with immunohistochemistry and A. Messer for assisting with the analysis of the canine samples. We also thank N. Hoepp for discussions on canine liver clinical pathology. We thank S. Sherrill-Mix for help with statistical analysis. This work was supported by grants from the National Institutes of Health (RO1HL083017 (H.H.K.), R24HL63098 and N0175N92019D00041 (T.C.N.), RO1HL126850 (D.E.S.) and RO1AI082020, RO1CA241762, RO1HL142791 and U19AI149680 (F.D.B)). We also acknowledge support from the Penn Center for AIDS Research (P30AI045008) and the PennCHOP Microbiome Program (F.D.B.).
Author information
Authors and Affiliations
Contributions
G.N.N., J.K.E., S.K., H.E.R., A.M.R., J.L. and C.W. performed the experiments. E.P.M., C.T.L. and T.C.N. performed the vector administration, sample collection and follow-up with the dogs. C.A.-A. performed the dog liver histopathology analysis. D.E.S., H.H.K., T.C.N. and F.D.B. designed the experiments. D.E.S. and F.D.B. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
D.E.S. receives royalties from a licensing agreement with Spark Therapeutics. D.E.S. and G.N.N. are inventors on a patent on FVIII and hemophilia A gene therapy.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
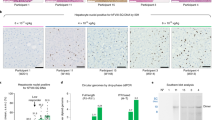
Extended Data Fig. 1 Immunohistochemical detection of FVIII in liver after AAV administration.
Immunohistochemical detection of FVIII in the liver from untreated (Hem A) (a) and treated (b,c,d,e,f) hemophilia A dogs. Locations of FVIII production are indicated by the brown stain. Most of the cFVIII staining was pan-lobular in distribution (b,c,d,e) while some areas had what appeared to be small clonal populations of cells that express cFVIII (b,f). Liver sections from multiple lobes were stained in n = 4 independent experiments. Images are representative of each dog. Scale bar representing 50μm applies to all images.
Supplementary information
Supplementary Information
Supplementary Figs. 1–12, Tables 1–12 and AAV Vector Sequences.
Rights and permissions
About this article
Cite this article
Nguyen, G.N., Everett, J.K., Kafle, S. et al. A long-term study of AAV gene therapy in dogs with hemophilia A identifies clonal expansions of transduced liver cells. Nat Biotechnol 39, 47–55 (2021). https://doi.org/10.1038/s41587-020-0741-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-020-0741-7
This article is cited by
-
Decoding the complexity of on-target integration: characterizing DNA insertions at the CRISPR-Cas9 targeted locus using nanopore sequencing
BMC Genomics (2024)
-
Improving CRISPR–Cas9 directed faithful transgene integration outcomes by reducing unwanted random DNA integration
Journal of Biomedical Science (2024)
-
Recent advances in various adeno-associated viruses (AAVs) as gene therapy agents in hepatocellular carcinoma
Virology Journal (2024)
-
Gene therapy corrects the neurological deficits of mice with sialidosis
Gene Therapy (2024)
-
Adeno-associated virus as a delivery vector for gene therapy of human diseases
Signal Transduction and Targeted Therapy (2024)