Abstract
Linker histone H1 proteins bind to nucleosomes and facilitate chromatin compaction1, although their biological functions are poorly understood. Mutations in the genes that encode H1 isoforms B–E (H1B, H1C, H1D and H1E; also known as H1-5, H1-2, H1-3 and H1-4, respectively) are highly recurrent in B cell lymphomas, but the pathogenic relevance of these mutations to cancer and the mechanisms that are involved are unknown. Here we show that lymphoma-associated H1 alleles are genetic driver mutations in lymphomas. Disruption of H1 function results in a profound architectural remodelling of the genome, which is characterized by large-scale yet focal shifts of chromatin from a compacted to a relaxed state. This decompaction drives distinct changes in epigenetic states, primarily owing to a gain of histone H3 dimethylation at lysine 36 (H3K36me2) and/or loss of repressive H3 trimethylation at lysine 27 (H3K27me3). These changes unlock the expression of stem cell genes that are normally silenced during early development. In mice, loss of H1c and H1e (also known as H1f2 and H1f4, respectively) conferred germinal centre B cells with enhanced fitness and self-renewal properties, ultimately leading to aggressive lymphomas with an increased repopulating potential. Collectively, our data indicate that H1 proteins are normally required to sequester early developmental genes into architecturally inaccessible genomic compartments. We also establish H1 as a bona fide tumour suppressor and show that mutations in H1 drive malignant transformation primarily through three-dimensional genome reorganization, which leads to epigenetic reprogramming and derepression of developmentally silenced genes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All sequencing data that support the findings of this study have been deposited in the NCBU Gene Expression Omnibus (GEO) with accession number GSE143293. All other data that support the findings of this study are available from the corresponding authors upon request. Source data are provided with this paper.
References
Fyodorov, D. V., Zhou, B. R., Skoultchi, A. I. & Bai, Y. Emerging roles of linker histones in regulating chromatin structure and function. Nat. Rev. Mol. Cell Biol. 19, 192–206 (2018).
Fan, Y. et al. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell 123, 1199–1212 (2005).
Cao, K. et al. High-resolution mapping of H1 linker histone variants in embryonic stem cells. PLoS Genet. 9, e1003417 (2013).
Zhang, Y. et al. Histone H1 depletion impairs embryonic stem cell differentiation. PLoS Genet. 8, e1002691 (2012).
Nacev, B. A. et al. The expanding landscape of ‘oncohistone’ mutations in human cancers. Nature 567, 473–478 (2019).
Bennett, R. L. et al. A mutation in histone H2B represents a new class of oncogenic driver. Cancer Discov. 9, 1438–1451 (2019).
Okosun, J. et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat. Genet. 46, 176–181 (2014).
Li, H. et al. Mutations in linker histone genes HIST1H1 B, C, D, and E; OCT2 (POU2F2); IRF8; and ARID1A underlying the pathogenesis of follicular lymphoma. Blood 123, 1487–1498 (2014).
Reichel, J. et al. Flow sorting and exome sequencing reveal the oncogenome of primary Hodgkin and Reed-Sternberg cells. Blood 125, 1061–1072 (2015).
Mesin, L., Ersching, J. & Victora, G. D. Germinal center B Cell dynamics. Immunity 45, 471–482 (2016).
Chapuy, B. et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat. Med. 24, 679–690 (2018).
Fan, Y. et al. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol. Cell. Biol. 23, 4559–4572 (2003).
Wright, G. W. et al. A probabilistic classification tool for genetic subtypes of diffuse large B cell lymphoma with therapeutic implications. Cancer Cell 37, 551–568 (2020).
Zhou, B. R. et al. Structural mechanisms of nucleosome recognition by linker histones. Mol. Cell 59, 628–638 (2015).
Lu, C. et al. Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science 352, 844–849 (2016).
Swaroop, A. et al. An activating mutation of the NSD2 histone methyltransferase drives oncogenic reprogramming in acute lymphocytic leukemia. Oncogene 38, 671–686 (2019).
Béguelin, W. et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell 23, 677–692 (2013).
Ortega-Molina, A. et al. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat. Med. 21, 1199–1208 (2015).
Eagen, K. P. Principles of chromosome architecture revealed by Hi-C. Trends Biochem. Sci. 43, 469–478 (2018).
Rao, S. S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Izzo, A. et al. The genomic landscape of the somatic linker histone subtypes H1.1 to H1.5 in human cells. Cell Rep. 3, 2142–2154 (2013).
Bascom, G. D., Myers, C. G. & Schlick, T. Mesoscale modeling reveals formation of an epigenetically driven HOXC gene hub. Proc. Natl Acad. Sci. USA 116, 4955–4962 (2019).
Kim, J. M. et al. Linker histone H1.2 establishes chromatin compaction and gene silencing through recognition of H3K27me3. Sci. Rep. 5, 16714 (2015).
Lhoumaud, P. et al. NSD2 overexpression drives clustered chromatin and transcriptional changes in a subset of insulated domains. Nat. Commun. 10, 4843 (2019).
Ezponda, T. et al. The histone methyltransferase MMSET/WHSC1 activates TWIST1 to promote an epithelial–mesenchymal transition and invasive properties of prostate cancer. Oncogene 32, 2882–2890 (2013).
Mulder, T. A., Wahlin, B. E., Österborg, A. & Palma, M. Targeting the immune microenvironment in lymphomas of B cell origin: from biology to clinical application. Cancers 11, 915 (2019).
Yang, S. M., Kim, B. J., Norwood Toro, L. & Skoultchi, A. I. H1 linker histone promotes epigenetic silencing by regulating both DNA methylation and histone H3 methylation. Proc. Natl Acad. Sci. USA 110, 1708–1713 (2013).
Geeven, G. et al. Local compartment changes and regulatory landscape alterations in histone H1-depleted cells. Genome Biol. 16, 289 (2015).
Wilcockson, M. et al. H1 histones control the epigenetic landscape by local chromatin compaction. Nature https://doi.org/10.1038/s41586-020-3032-z (2020).
Eminli, S. et al. Differentiation stage determines potential of hematopoietic cells for reprogramming into induced pluripotent stem cells. Nat. Genet. 41, 968–976 (2009).
Wong, D. J. et al. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell 2, 333–344 (2008).
Stadhouders, R. et al. Transcription factors orchestrate dynamic interplay between genome topology and gene regulation during cell reprogramming. Nat. Genet. 50, 238–249 (2018).
Acknowledgements
E.C., A.M.M. and C.D.A. are funded through NIH/NCI R01 CA234561 and STARR I9-A9-062. A.M.M. and A.T. are funded by NIH/NCI P01 CA229086. Research in the C.D.A. laboratory is also supported by the NCI P01 CA196539 Leukemia and Lymphoma Society (LLS-SCOR 7006-13), The Rockefeller University and St Jude Children’s Research Hospital Collaborative on Chromatin Regulation in Pediatric Cancer and Robertson Therapeutic Development Fund. A.M.M. is also funded by NIH/NCI R35 CA220499, LLS TRP 6572, LLS SCOR 7012, the Follicular Lymphoma Consortium, the Samuel Waxman Cancer Research Foundation and the Chemotherapy Foundation. J.D.L., A.M.M. and C.D.A. are funded by LLS SCOR 17403-19 and J.D.L. is funded by R01 CA195732 and The Samuel Waxman Cancer Research Foundation. N.Y. is funded by the Congressionally Directed Medical Research Program (CA181397). Research in EpiCypher is supported by R44 DE029633 and R44 GM116584. A.A.S. was funded by the Damon Runyon Cancer Research Foundation (DRG-2185-14). A.I.S. is funded through GM116143. Histone proteomics work was performed at Northwestern Proteomics, which was supported by NCI CCSG P30 CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center, an instrumentation award (S10OD025194) from the NIH Office of Director, and the National Resource for Translational and Developmental Proteomics supported by P41 GM108569. The in silico modelling work was supported by award R35-GM122562 to T.S. The authors thank the Laboratory of Comparative Pathology, Epigenomics Core, Flow Cytometry Core Facility and Optical Microscopy Core at Weill Cornell Medicine; the Genomics Resource Center and Bio-Imaging Resource Center at The Rockefeller University; and Langone Health’s Genome Technology Center at New York University.
Author information
Authors and Affiliations
Contributions
N.Y., A.A.S., E.C. and A.M.M conceived and designed the study and wrote the manuscript. N.Y. performed in vivo experiments. A.A.S., J.M.C. and A.O. performed biochemical experiments. A.K., M.T., C.R.C., C.E.M., A.S.D. and M.I. performed bioinformatics analysis on sequencing-based data. N.Y., A.A.S., A.O., O.E., J.M.P., E.A., Y.D., N.L.K., A.T. and L.M.S. analysed data. J.C. performed mouse genotyping and breeding. B.J.V. and M.-C.K. provided CUT&RUN data. D.W.S. and C.D.A. provided resources. W.B. and E.A. were involved in designing experiments. J.D.L. analysed data and helped write the manuscript. A.I.S. provided the H1 knockout mouse model. T.S. and S.P.-L. performed the in silico modelling. All authors contributed to the written manuscript.
Corresponding authors
Ethics declarations
Competing interests
A.M.M. has research funding from Janssen Pharmaceuticals, Sanofi and Daiichi Sankyo, has consulted for Epizyme and Constellation and is on the advisory board for KDAC Pharma. A.M.M. has also been on advisory boards for BMS and ExoTherapeutics. N.L.K. is a consultant for Thermo Fisher Scientific. C.D.A is a co-founder of Chroma Therapeutics and Constellation Pharmaceuticals, and a Scientific Advisory Board member of EpiCypher. EpiCypher is a commercial developer of the CUTANA CUT&RUN platform. M.I. has received consultancy fees from Novartis Venture Fund outside of the scope of the work. The remaining authors declare no competing interests.
Additional information
Peer review information Nature thanks Iannis Aifantis, Ralph Stadhouders and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Characterization of H1 allele mutations and deletions in DLBCL.
a, A PanCancer Atlas cancer mutation survey was performed using the cBioPortal to search the cumulative mutation frequency of H1A–H1E across a total of 10,953 non-redundant patient samples across all cancer types. b, H1B–H1E mutation landscape across non-redundant TCGA and BCCA (British Columbia Cancer Agency) DLBCL samples (number of samples indicated for each H1 isoform); location of mutations is plotted onto protein structure, with bar height corresponding to mutation counts, total frequency of mutations in each H1 isoform is plotted to the right (red bars). c, Prevalence of mutant H1 versus wild-type H1 cases in ABC and GC B DLBCLs (Fisher’s exact test for enrichment in ABC or GC B-DLBCL, P > 0.05) d, Prevalence of H1A–H1E missense mutations and heterozygous loss in MCD-subtype DLBCL and their enrichment (−log10(P value)) over non-MCD DLBCLs. e, Summary table of frequency of mutations and heterozygous loss of H1A–H1E in DLBCL subtypes (MCD, ST2, BN2, EZB, N1 and AP53). f, Co-occurrence as odds ratio and –log10(P value) among H1 alleles in 101 germline-matched WGS DLBCLs. g, Frequency and location of missense mutations for H1A–H1E in 101 germline-controlled DLBCLs. h, Quantile-quantile plot showing the P values for SNVs across 101 germline-matched WGS DLBCLs. Driver analysis derived as probability of mutation count greater than or equal to the observed mutation count under P values expected from a Gamma-Poisson distribution for SNVs. Shaded grey zone contains mutant genes with FDR < 0.01, depicted as blue or red dots, several of which are listed in the figure. i, Oncoprint for H1A–H1E in 101 germline-matched WGS DLBCLs. j, Co-occurrence as odds ratio (OR) and –log10(P value) among H1 alleles in 101 germline-matched WGS DLBCLs.
Extended Data Fig. 2 H1 mutations are genetic drivers in lymphoma and confer loss of function.
a, Crystal structure of the linker histone globular domain (grey) bound to nucleosome (Protein Data Bank (PDB), 4QLC), with magnified view of ASGS amino acid residues highlighted in red. b, Representative images of FRAP of ectopically expressed, monomeric eGFP-tagged wild-type H1C and H1C mutants S102F and P118S in 3T3 cells before, immediately after, and at 10, 30 and 90 s after bleaching the area (yellow dashed square). Scale bars, 5 μm. c, Quantification of normalized intensity as representation of turnover kinetics from b for wild-type H1C (n = 18) and mutants A101V (n = 15), S102F (n = 9), S104F (n = 10), G103A (n = 10) and P118S (n = 10); shaded area indicates 95% confidence intervals. Data are pooled from two independent biological experiments. d, Dissociation constant (Kd) of the binding to mononucleosomes of recombinant H1C mutants S102F and P118S compared to wild-type H1C, as determined by biolayer interferometry. Data are mean ± s.e.m. (two-sided unpaired t-test, ****P < 0.0001). Data are global fits from five concentration measurements. e, Chromatin fibre oligomerization upon serial precipitation by Mg2+ as per cent soluble 12-mer arrays was determined for no H1, wild-type H1C and the C-terminal P118S mutant. Data are mean ± s.d. Data are pooled from three independent biological experiments. f, Atomic force microscopy imaging of chromatin arrays in the presence of wild-type H1 and the C-terminal P118S mutant. Scale bars, 200 nm. Images are representative of two independent biological experiments.
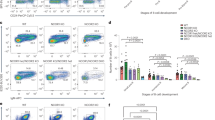
Extended Data Fig. 3 H1c−/−H1e−/− mature B cells show normal development in spleen and bone marrow.
a, mRNA of human H1B–H1E normalized to RPL13A in GC B cells relative to naive B cells (H1B, **P = 0.004; H1E, *P = 0.027), isolated from three independent specimens of human tonsils. Data are mean ± s.d., two-sided unpaired t-tests. b, Mouse H1b–H1e mRNA levels normalized to Gapdh in sorted GC B cells (n = 3) relative to naive B cells(n = 3) (H1b, ****P < 0.0001). Data are mean ± s.d., two-sided unpaired t-tests. c, Quantification of spleen/body weight ratios of two-month-old H1c−/−H1e−/− (n = 13) and wild-type littermate control (n = 14) mice. Data are pooled from two independent experiments. P < 0.05; NS, not significant; two-sided unpaired t-tests. Data are mean ± s.d. d, Quantification of GC area (Ki67 staining) in the spleens of H1c−/−H1e−/− (n = 10) and WT (n = 10) mice. ***P = 0.0005. Data are mean ± s.d., two-sided unpaired t-tests. e, f, Immunohistochemistry images of spleen sections of cleaved caspase-3 (e) and γ-H2AX (f) staining (left) and quantification (right) of positively stained follicular cells from H1c−/−H1e−/− (n = 3) and wild-type littermate control (n = 3) mice immunized with SRBCs and euthanized 10 days after immunization. Scale bars, 100 μm. P < 0.05 (not significant; NS), two-sided unpaired t-tests. Data are mean ± s.d. g, Flow cytometry analysis and quantification of (Fas+CD38−) GC B cells within total B cells from H1c−/−H1e−/− and wild-type mice (n = 10 per genotype). **P = 0.0018, two-sided unpaired t-tests. Data are mean ± s.d. h, Quantification of the percentage of B220+ splenocytes in H1c−/−H1e−/− (n = 10) and wild-type (n = 10) mice 9 days after SRBC immunization. P < 0.05, two-sided unpaired t-tests. Data are mean ± s.d. i, Flow cytometry analysis and quantification of GC B cells (Fas+GL7+) from H1c−/−H1e−/− (n = 10) and wild-type (n = 10) mice. Two-sided unpaired t-tests, *P = 0.041. Data are mean ± s.d. j, Flow cytometry analysis and quantification of mature B cells (B220+IgD+IgM+) and transitional B cells (B220+IgDintIgM+) in spleens from H1c−/−H1e−/− (n = 10) and wild-type (n = 10) mice. P < 0.05, two-sided unpaired t-tests. Data are mean ± s.d. k, Flow cytometry quantification of follicular B cells (B220+D23+CD21+) and marginal zone B cells (B220+D23loCD21+) in spleens from H1c−/−H1e−/− (n = 10) and wild-type (n = 10) mice. P < 0.05, two-sided unpaired t-tests. Data are mean ± s.d. l, Flow cytometry analysis gated on B220+CD24+ and quantification of ProBPreB (IgM−IgD−), immature (IgM−IgDlo), transitional (IgD+IgM−) and early mature (IgD+IgM+) B cells in bone marrow of H1c−/−H1e−/− (n = 4) and wild-type (n = 5) mice. P < 0.05, two-sided unpaired t-tests. Data are mean ± s.d. m, Percentage of Ki67+early B cells (B220+CD24+) in bone-marrow of H1c−/−H1e−/− (n = 4) and wild-type (n = 5) mice, as well as naive B cells (***P = 0.0004)and marginal zone B cells (***P = 0.001) in the spleens of H1c−/−H1e−/− (n = 5) and wild-type (n = 5) mice. n, Schematic diagram of primary immunization with NP-KLH and secondary immunization 21 days after with NP-CGG. o, Ratio between high (NP8) and low (NP30) affinity NP-specific IgG1 antibody titres in sera of H1c−/−H1e−/− (n = 5) and wild-type (n = 5) mice by enzyme-linked immunosorbent assay (ELISA). P < 0.05, two-sided unpaired t-tests. Data are mean ± s.d. p, Enzyme-linked immunosorbent spot (ELISPOT) quantification of NP-specific (anti-NP8 and anti-NP30) IgG1-secreting cells from the bone marrow of H1c−/−H1e−/− (n = 5) and wild-type (n = 5) mice. P < 0.05, two-sided unpaired t-test. Data are mean ± s.d. Data are representative of two independent experiments. q, Representative images of anti-NP8 and anti-NP30 96-well ELISPOT. r, Flow cytometry analysis and quantification of centroblasts within the dark zone (DZ) (CXCR4+CD86−) (***P = 0.0002) and centrocytes within the light zone (LZ) (CXCR4−CD86+) (***P = 0.0002) within GC B cells from H1c−/−H1e−/− (n = 10) and wild-type (n = 10) mice. Two-sided unpaired t-test. Data are representative of three independent experiments. s, Immunofluorescence confocal microscopy images of GCs at day 7 after immunization in mixed chimaeras. Scale bar, 50 μm. Images are representative of two independent experiments. t, Quantification from s of the fraction of PNA+CD45.1 or CD45.2 cells (17 GCs, n = 3 mice). Two-sided paired t-test, ***P = 0.0004. u, Relative EdU+ GC B cell/GC B cell fraction for wild-type CD45.1+and H1c−/−H1e−/− CD45.2+ cells at day 7 after immunization (n = 4 chimaeras). Two-sided paired t-test, **P = 0.0065. Data are representative of two independent experiments.
Extended Data Fig. 4 Loss of H1c and H1e induces stem-cell-like transcriptional profiles in GC B cells and DLBCLs.
a, Unsupervised hierarchical clustering analysis of RNA-seq data from sorted H1c−/−H1e−/− and wild-type H1 GC B cells, based on genes in the top 90th percentile of variability. b, Expression of NSD2 (measured as fragments per kilobase of exon model per million reads mapped; FPKM) from RNA-seq profiles of human and mouse naive B and GC B cells. c, GSEA analysis of genes linked to NSD2 gain-of-function mutation in three cell lines (RCHACV, SEM, RPMI) against ranked gene expression changes in mouse H1c−/−H1e−/− GC B cells. d, Box plot of log2-transformed relative gene expression normalized to the average expression value of all genes from the top 200 differentially upregulated genes in the H1c−/−H1e−/− signature against the ImmGen database. Box plot centre represents median, bounds of box are the first and third quartiles and whiskers extend to 1.5 × the interquartile range. e, GSEA analysis with indicated gene sets, using ranked log2-transformed fold change in expression between H1c−/−H1e−/− and wild-type mouse GC B cells. f, GSEA analysis of gene sets linked to EZH2 against ranked gene expression changes in mouse H1c−/−H1e−/− GC B cells. g, Volcano plot showing differentially expressed genes between patients with H1C/E-mutant DLCBL (n = 18) versus patients with H1-wild-type DLBCL (n = 237) (FDR < 0.05, fold change > 1.5). Red field denotes upregulated and blue field downregulated genes. h, GSEA analysis of upregulated genes in patients with H1C/E -mutant versus H1-wild-type DLBCL, using ranked changes in log2-transformed fold change in mouse H1c−/−H1e−/− GC B cells. i, Gene pathway enrichment analysis of upregulated and downregulated genes from g, hypergeometric mean test. j, k, Sorted H1c−/−H1e−/− or wild-type GC B cells (n = 2 per genotype) were subjected to droplet based (10X) single-cell RNA-seq. Centroblasts (j) and centrocytes (k) were defined on the basis of enrichment for centroblast and centrocyte signature profiles, respectively projected onto the uniform manifold approximation and projection (UMAP) distribution of cells. l, Top, expression of G2M cell cycle proliferation gene signature was plotted for each cell on the y axis with spline curves representing the average for H1c−/−H1e−/− and wild-type GC B cells. Bottom, differential expression is shown as a delta spline plot (blue) across pseudotime and tested by two-sided Wilcoxon test within ten bins of equal cell number (dashed lines).
Extended Data Fig. 5 HiC compartment analysis and integration with ATAC–seq in H1c−/−H1e−/− and wild-type GC B cells.
a, Genome-wide correlation score (Stratum adjusted correlation score, SCC) of HiC matrices within same genotype (***P = 0.0002171, two-sided unpaired t-test), and across genotypes (P < 0.05, two-sided Wilcoxon test). b, Principal component analysis (PCA) of compartment bins processed with Hi-C bench at a resolution of 10 0kb from Hi-C replicates for H1c−/−H1e−/− and wild-type GC B cells. c, Volcano plot showing significant c-score shifts in H1c−/−H1e−/− mouse GC B cells based on Δ c-score and −log10(permutation FDR P value): decompacted (red) compartment bins (n = 5,320) and compacted (blue) compartment bins (n = 386). d, Δ c-score (H1c−/−H1e−/− versus wild type) across mouse chromosomes (positive y axis is in red for decompacting loci and negative y axis is blue for compacting loci) plotted as ideograms. e, Box plots of Δ c-score (H1c−/−H1e−/− versus wild type) across c-score range (−1.0 to 1.0) separated into 0.1 bins. No statistical evaluation was derived for this graph. f, Volcano plot showing TADs with significant gain of intra-TAD interactivity (n = 26, red) and reduction of intra-TAD interactivity (n = 2, blue; two-sided unpaired t-test, FDR-adjusted P value < 0.05, fold change > 1.5). g, Volcano plot showing significant ATAC–seq peaks gaining accessibility (n = 438) and losing accessibility (n = 53; two-sided unpaired t-test FDR-adjusted P value < 0.05, fold-change > 1.5). h, Scatter plot showing log2-transformed fold change in ATAC–seq peaks between H1c−/−H1e−/− and wild-type GC B cells in decompacting compartments versus wild-type c-score: B to BwA (left), B to A (middle), and A to AwA (right). Peaks gaining accessibility (FC > 1.5, Padj < 0.05) are marked in red. Estimated odds ratios and P values were calculated using Fisher’s exact test. Although ATAC–seq peaks are more prevalent in the A-to-AwA compartment (right), increased accessibility is more enriched in B-to-BwA and B-to-A compartments. i, HiC contact maps of regions surrounding Spry (left), Tusc1 (middle) and Meis1(right) genes. The top of each square shows H1 wild-type contacts and the bottom of each square shows those in H1c−/−H1e−/− GC B cells. Heat maps represent the Pearson correlation of interactions in wild-type and H1c−/−H1e−/− GC B cells. Bottom tracks represent the eigenvector (PC1) for compartments A and B in red and blue, respectively, and show the position of genes within these loci. j, k, GSEA analysis of genes shifting to A compartments (B to A, B to BwA, A to AwA) or genes contained in stable compartments using ranked log2-transformed gene expression in H1c−/−H1e−/− GC B cells. (NES and FDR values as implemented by GSEA). l, Gene pathway enrichment analysis of genes in decompacting and stable compartments in H1c−/−H1e−/− versus wild-type GC B cells (hypergeometric mean test).
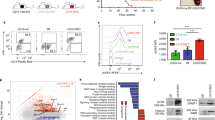
Extended Data Fig. 6 Three-dimensional changes in chromatin architecture owing to a deficiency of H1C and H1E in GC B cells recapitulate decompaction during iPS cell differentiation.
a, Schematic of a previous study of B cell differentiation to iPS cells32 with time points for HiC analysis. b, Correlation plots comparing shifting to A c-scores in iPS cells (day 2, day 4, day 6, day 8) compared to control B cells versus c-score changes in H1c−/−H1e−/− GC B cells. Estimated odds ratios and P values were calculated using Fisher’s exact test. c, GSEA analysis of shifting to A compartments in iPS cells (day 2, day 4, day 6, day 8) against ranked Δ c-scores derived from H1c−/−H1e−/− minus wild-type mouse GC B cells (NES and FDR values as implemented by GSEA). d, Violin plots comparing stable and shifting B to A compartments during iPS cell differentiation (day 2, day 4, day 6, day 8 and fully undifferentiated) to the estimated Δ c-score due to deficiency of H1C and H1E in those same regions in GC B cells. (day 2, P < 2.2 × 10−16; day 4, P < 2.2 × 10−16; day 6, P = 0.002; day 8, 0.0001; estimated with two-sided Wilcoxon test). Box plot centre represents median, bounds of box are the first and third quartiles and whiskers extend to 1.5 × the interquartile range. e, v4c analyses on the Klf5 locus (chr14: 99,000,000–100,200,000) anchored on the Klf5 promoter for (top) B cell reprogramming (blue) to iPS cells (red) with four intermediate time states (grey) from a previous study32, as well as GC B cells (bottom) (H1c−/−H1e−/− and wild-type H1). IgV tracks below comprise Δ c-score and ATAC–seq signal in H1c−/−H1e−/− versus wild-type GC B cells. Gained HiC interactions in H1c−/−H1e−/− compared to wild-type GC B cells (shaded in grey 1–3: pval = 0.04; pval = 0.059; pval = 0.02, respectively, two-sided unpaired t-test) have OCT2 motif sequences as shown. f, Schematic of experimental set-up with H1c−/−H1e−/− or wild-type littermate mouse embryonic fibroblasts. g, Representative images of alkaline phosphatase (AP)-stained H1c−/−H1e−/− and wild-type iPS cell colonies at day 21. h, Per cent iPS cell reprogramming efficiency of H1c−/−H1e−/− (n = 5 transfections on two biological replicates) and wild-type (n = 4 transfections on two biological replicates) mouse embryonic fibroblasts determined as the ratio of AP+ colonies to the number of seeded mCherry+ cells. P = 0.01, two-sided unpaired t-tests. Data are mean ± s.d. Data are representative of three independent experiments.
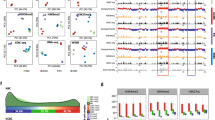
Extended Data Fig. 7 Altered H3K36me2 and H3K27me3 distribution in H1c−/−H1e−/− GC B cells.
a, b, Mass spectrometry of H3K36 (a) and H3K27 (b) post-translational modifications, log2-transformed and normalized to average wild-type peak area from H1c−/−H1e−/− (n = 5) and wild-type (n = 7) acid-extracted samples from GC B cells; two-sided unpaired t-test: K36 unmodified (unmod.), ***P = 0.0005; K36me1, ***P = 0.0003; K36me2 ***P = 0.0002; K36me3, P = 0.93; K36ac, P = 0.56; K27 unmod., *P = 0.0157; K27me1, **P = 0.0072; K27me2, *P = 0.0175; K27me3, ***P = 0.0007; K27ac, P = 0.9337. Box plots show median and 25th to 75th percentiles, whiskers indicate data range. Data are representative of two independent experiments. c, Immunoblots for H1 (D4J5Q and AE-4 antibodies), H3K36me2, H3K27me3, EZH2 and NSD2 from sorted wild-type and H1c−/−H1e−/− GC B cells. Direct blue stain is included as loading control. A representative image of at least three experiments is shown. Uncropped gels are shown in Supplementary Fig. 1. d, Mass-spectometry-based relative abundance of H3.1 or H3.2 (replication-dependent) and H3.3 (replication-independent) isoforms, shown as the average percentage of the total peak area of H3K27-K36 peptide containing H3.3-specific Ser31 in acid-extracted histones from wild-type (H3.3, 15.76%, n = 7) and H1c−/−H1e−/− (H3.3, 15.07%, n = 5) GC B cells; two-sided unpaired t-test, P = 0.0004, Data are mean ± s.d. e, Mass spectrometry of H3 K36 (top) and K27 (bottom) post-translational modifications across H3.1/2 (left) and H3.3 (right) isoforms, log2-transformed and normalized to average wild-type peak area from samples acid-extracted from wild type (n = 7) and H1c−/−H1e−/− (n = 5) GC B cells; two-sided unpaired t-test: H3.1/2 K36 unmod., ***P = 0.0005; H3.1/2 K36me1, ***P = 0.0003; H3.1/2 K36me2, ****P < 0.0001; H3.1/2 K36me2, P = 0.88; H3.1/2 K27 unmod., *P = 0.0100; H3.1/2 K27me1, *P = 0.0162; H3.1/2 K27me2, *P = 0.0129; H3.1/2 K27me3, ***P = 0.0002; H3.3 K36 unmod., ****P < 0.0001; H3.3 K36me1, *P = 0.036; H3.3 K36me2, P = 0.15; H3.3 K36me3, P = 0.5974; H3.3 K27 unmod., P = 0.1187; H3.3 K27me1, P = 0.4743; H3.3 K27me2, P = 0.1199; H3.3 K27me3, P = 0.0628. Box plots show median and 25th to 75th percentiles, whiskers indicate data range. f, Unsupervised hierarchical clustering analysis of ChIP–seq data for H3K27me3 and H36me2 in biological triplicates from sorted H1c−/−H1e−/− and wild-type GC B cells. g, Genome-wide correlation plot of log2-transformed fold change (H1c−/−H1e−/− versus wild type) of normalized reads within ChIP–seq peak union for H3K36me2 and H3K27me3 (Pearson correlation coefficient R = −0.453, P < 1 ×10−16). h, Heat map of HiC c-score, H3K36me2 and H3K27me3 centred within shifting B-to-A compartments (100 kb) and surrounding 300 kb for H1c−/−H1e−/− and wild-type GC B cells for compartment ‘extensions’ (top). i, Fraction of ChIP–seq peak coverage (H3K36me2 in red and H3K27me3 in blue) within 100-kb compartments across HiC c-score (x axis, −1 to 1) for wild-type GC B cells. Cubic smoothing splines of data are presented with shaded regions indicating 99% confidence intervals. j, Fraction peak (H3K27me3 and H3K36me2) coverage of regions within shifting compartment groups 1–5 in H1c−/−H1e−/− and wild-type GC B cells. Paired Wilcoxon test, group 1: H3K27me3, P < 1 ×10−16; H3K36me2, P = 0.597; group 2: H3K27me3, P < 1 ×10−16; H3K36me2, P < 1 ×10−16; group 3: H3K27me3, P = 3.38 ×10−13; H3K36me2, P < 1 ×10−16; group 4: H3K27me3, P = 8.71 ×10−15; H3K36me2, P < 1 ×10−16; group 5: H3K27me3, P < 1 ×10−16; H3K36me2, P < 1 ×10−16. Box plot centre represents median, bounds of box are the first and third quartiles and whiskers extend to 1.5 × the interquartile range. k, Scatter plot of H3K27me3 peak log2-transformed fold change (H1c−/−H1e−/− versus wild-type GC B cells) versus wild-type c-score score for decompacting group 2. Gain of H3K27me3 (red dots) largely occurred within regions shifting from compartment B whereas loss of H3K72me3 (blue dots) was more prevalent within regions shifting compartments from compartment A.
Extended Data Fig. 8 Changes in activation marks H3K4me3 and H3K27ac and repressive marks H3K9me2 and H3K9me3 in H1-deficient GC B cells are associated with compartment-B decompaction.
a, Genes defined as groups 3 and 4 (n = 108 genes) and group 5 (n = 152 genes) show transcriptional activation and significant upregulation in H1c−/−H1e−/− GC B cells compared to wild-type GC B cells. Paired Wilcoxon test. Box plot centre represents median, bounds of box are the first and third quartiles and whiskers extend to 1.5 × the interquartile range. b, Top, expression of the genes defined as groups 3–5 was plotted for each cell on the y axis. Average expression is represented by the different coloured spline curves for each genotype as indicated. Bottom, differential expression between H1c−/−H1e−/− and wild-type cells is represented as the delta spline plot across pseudotime. Cells are divided by pseudotime into bins of equal cell number (grey vertical dashed lines) and tested for signature enrichment compared to wild-type GC B cells by two-sided Wilcoxon test (P < 6.81 × 10−50) c, GSEA with genes marked with gain in H3K4me3 (top) or H3K27Ac (bottom) on their promoters (transcription start site (TSS) ± 500 bp) using the ranked log2-transformed fold change in mouse H1c−/−H1e−/− GC B cells. d, Fraction of histone peak coverage for H3K4me3 and H3K27ac within 100-kb compartments across HiC c-score (x axis) for wild-type GC B cells. Cubic smoothing splines of data are presented with shaded regions indicating 99% confidence intervals. e, Fraction of histone peak coverage (for H3K9me2 and H3K9me3) within 100-kb compartments across HiC c-score (x axis) for wild-type GC B cells. Cubic smoothing splines of data are presented with shaded regions indicating 99% confidence intervals. f, CUT&RUN peaks for H3K9me2 and H3K9me3 show altered abundance between H1c−/−H1e−/− and wild-type GC B cells (FC > 1.5, n = 5,277 gained and n = 4,717 lost H3K9me2 and n = 2,511 gained and n = 7,529 lost H3K9me3 peaks). g, Genome-wide correlation plot of log2-transformed fold change (H1c−/−H1e−/− versus wild-type H1 GC B cells) in H3K9me2 (left) and H3K9me3 (right) peaks versus Δ c-score (H1c−/−H1e−/− minus wild-type H1). Both H3K9me2 and H3K9me3 changes were largely loss and were anti-correlated with compartment decompaction (Pearson correlation coefficient R = −0.438, P < 1 ×10−16 and R = −0.543, P < 1 ×10−16, respectively). h, i, Heat maps of H39me2 and H3k9me3 centred within shifting B-to-A compartments (100 kb) and surrounding 300 kb for H1c−/−H1e−/− and wild-type GC B cells for compartment ‘extensions’ (h) and ‘islands’ (i). j, Fraction peak (H3K9me2 and H3K9me3) coverage of regions within shifting compartment groups and 2 in H1c−/−H1e−/− and wild-type GC B cells. Paired Wilcoxon test, group 1: H3K9me2, P < 1 ×10−16, H3K9me3, P < 1 ×10−16; group 2: H3K9me2, P < 1 ×10−16; H3K9me3, P < 1 ×10−16. Box plot centre represents median, bounds of box are the first and third quartiles and whiskers extend to 1.5 × the interquartile range.
Extended Data Fig. 9 Linker histone incorporation reduces interactivity of chromatin fibre.
a, Representative equilibrated configurations of 50-nucleosome chromatin fibres obtained in silico in the absence of H1 and in the presence of 0.25, 0.5, 0.75 and 1 H1 molecule per nucleosome (H1C (left) and H1E (right)). Fibre contour (in red) on which the H1 mean positions are shown is shown on the top right of each model. Colour key shows DNA, linker histone, H2A, H2B, H3 and H4 tails. b, c, Contact maps for the 1,000-configuration ensembles obtained from left to right, in the absence of H1 and in the presence of 0.25, 0.50, 0.75 and 1 H1 per nucleosome(b, H1C; c, H1E). d, The nucleosome–nucleosome interaction patterns, or a one-dimensional decomposition of each contact map shown in b and c, indicate the dominant zigzag pattern of the fibre (i ± 2) and increase of long-range interactions as the density of H1C (left) or H1E (right) decreases. e, Packing ratio calculated as the number of nucleosomes contained in 11 nm of fibre for systems without H1 and in the presence of 0.25, 0.50, 0.75 and 1 H1 molecule per nucleosome (left, H1C; right, H1E); n = 1,000 chromatin ensemble configurations for each H1 per nucleosome ratio. Ordinary one-way analysis of variance for multiple comparisons, ****P < 0.0001. f, Volume of chromatin fibres calculated assuming a cylindrical shape for systems with no H1 and in the presence of 0.25, 0.5, 0.75 and 1 H1 molecule per nucleosome (left, H1C; right, H1E); n = 1,000 chromatin ensemble configurations for each H1 per nucleosome ratio. Ordinary one-way analysis of variance for multiple comparisons, ****P < 0.0001. Box plot centre represents median, bounds of box are the first and third quartiles and whiskers extend to 1.5 × the interquartile range (e, f).
Extended Data Fig. 10 Loss of H1c and H1e leads to aggressive Vav-PBcl2 lymphomas with DLBCL-like morphology.
a, Immunohistochemistry images of lymph node stained for H&E and B220 from VavP-Bcl2;H1c−/−H1e−/−, VavP-Bcl2;H1c−/+H1e−/+ and VavP-Bcl2-only mice at day 164. Scale bar, 1 mm. Images are representative of n = 11 mice per genotype examined over two independent experiments. b, Representative immunohistochemistry images of lymphomatous VavP-Bcl2;H1c−/−H1e−/−, VavP-Bcl2;H1c−/+H1e−/+and VavP-Bcl2-only lymph nodes stained for H3K36me2, and quantification of intensity (binned as high, mid, low and negative). Scale bar, 50 μm. Tissue derived from three mice per genotype with four tumour lymph nodes each. Data are mean ± s.d., two-sided unpaired t-test. c, Immunohistochemistry images of lung tissue stained for H&E, B220 and Ki67 from VavP-Bcl2;H1c−/−H1e−/−, VavP-Bcl2;H1c−/+H1e−/+ and VavP-Bcl2-only mice at day 164. Scale bar, 100 μm. Images are representative of n = 11 mice per genotype examined over two independent experiments. d, Quantification of B220+ lesion areas in liver tissue (Fig. 5c) from VavP-Bcl2;H1c−/−H1e−/− (****P < 0.0001) and VavP-Bcl2;H1c−/+H1e−/+(*P = 0.0308) compared to VavP-Bcl2-only (n = 7 mice per genotype, mean ± s.d.; two-sided unpaired t-tests). e, Immunohistochemistry stains for CD3 from VavP-Bcl2;H1c−/−H1e−/−and VavP-Bcl2;H1c−/+H1e−/+ lymphomas. Scale bar, 50 μm. Images are representative of n = 4 mice per genotype examined over two independent experiments. f, PCR for Igλ clonal rearrangement to report on tumour clonality of B220+ cells from VavP-Bcl2;H1c−/−H1e−/−, VavP-Bcl2;H1c−/+H1e−/+ and VavP-Bcl2-only mice at day 164. g, Immunohistochemistry images of liver and lung tissues stained for H&E and B220 in from H1c−/−H1e−/−, H1c−/+H1e−/+ and wild-type mice at day 164. Scale bar, 500 μm. Images are representative of n = 6 mice per genotype examined over two independent experiments. h, Immunohistochemistry images of lymph node tissue stained for H&E and B220 from VavP-Bcl2;H1c−/+H1e−/+ and VavP-Bcl2-only mice. Images are representative of n = 4 mice per genotype examined over two independent experiments. Scale bars, 100 μm. i, GSEA with the VavP-Bcl2;H1c−/−H1e−/− versus VavP-Bcl2 lymphoma upregulated gene set ranked against log2-transformed changes in fold change from mouse VavP-Bcl2;H1c−/+H1e−/+ versus VavP-Bcl2. j, Top, GSEA for genes upregulated in VavP-Bcl2;H1c−/+H1e−/+ versus VavP-Bcl2 lymphomas using the ranked log2-transformed fold change in mouse H1c−/−H1e−/− GC B cells. Bottom, GSEA for genes upregulated in VavP-Bcl2;H1c−/−H1e−/− versus VavP-Bcl2 mice using the ranked log2-transformed fold change in mouse H1c−/−H1e−/− GC B cells. k, Heat map showing differential expression of leading-edge genes (n = 898) from VavP-Bcl2;H1c−/−H1e−/− and VavP-Bcl2;H1c−/+H1e−/+ lymphomas. l, GSEA for genes upregulated in human H1C/E-mutant DLBCL, against the ranked log2-transformed fold change gene expression profiles of mouse VavP-Bcl2;H1c−/+H1e−/+ (left) and VavP-Bcl2;H1c−/−H1e−/− (right) lymphomas. m, Summary model depicting chromatin as contiguous B-to-A space, with H3K27 and H3K36 methylations occupying distinct compartments within. Loss of H1 results in a global shift of compartment interactivity towards A, with both H3K27 and H3K36 methylations shifting into ectopic regions. Most compacted regions devoid of either K27 or K36 methylation appear largely protected from H1 loss. The biological effects of H1 loss in GC B cells are summarized below.
Supplementary information
Supplementary Information
This file contains Supplementary Figure 1: Uncropped gel blots and Supplementary Figure 2: Flow cytometry and sorting gating strategies.
Supplementary Information
This file contains Supplementary Methods, which includes Supplementary Tables and Figures.
Rights and permissions
About this article
Cite this article
Yusufova, N., Kloetgen, A., Teater, M. et al. Histone H1 loss drives lymphoma by disrupting 3D chromatin architecture. Nature 589, 299–305 (2021). https://doi.org/10.1038/s41586-020-3017-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-3017-y
This article is cited by
-
Histone H1.0 couples cellular mechanical behaviors to chromatin structure
Nature Cardiovascular Research (2024)
-
Nucleosome reorganisation in breast cancer tissues
Clinical Epigenetics (2024)
-
NSD2 E1099K drives relapse in pediatric acute lymphoblastic leukemia by disrupting 3D chromatin organization
Genome Biology (2023)
-
Linker histone variant H1.2 is a brake on white adipose tissue browning
Nature Communications (2023)
-
EZH1/2 as targets for cancer therapy
Cancer Gene Therapy (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.