Abstract
The neural crest, an embryonic stem-cell population, is a vertebrate innovation that has been proposed to be a key component of the ‘new head’, which imbued vertebrates with predatory behaviour1,2. Here, to investigate how the evolution of neural crest cells affected the vertebrate body plan, we examined the molecular circuits that control neural crest development along the anteroposterior axis of a jawless vertebrate, the sea lamprey. Gene expression analysis showed that the cranial subpopulation of the neural crest of the lamprey lacks most components of a transcriptional circuit that is specific to the cranial neural crest in amniotes and confers the ability to form craniofacial cartilage onto non-cranial neural crest subpopulations3. Consistent with this, hierarchical clustering analysis revealed that the transcriptional profile of the lamprey cranial neural crest is more similar to the trunk neural crest of amniotes. Notably, analysis of the cranial neural crest in little skate and zebrafish embryos demonstrated that the transcriptional circuit that is specific to the cranial neural crest emerged via the gradual addition of network components to the neural crest of gnathostomes, which subsequently became restricted to the cephalic region. Our results indicate that the ancestral neural crest at the base of the vertebrate lineage possessed a trunk-like identity. We propose that the emergence of the cranial neural crest, by progressive assembly of an axial-specific regulatory circuit, allowed the elaboration of the new head during vertebrate evolution.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All raw sequencing data for all RNA-seq libraries (Fig. 3) and merged reference transcriptomes are available online (NCBI BioProject# PRJNA497902). Sequences of in situ probe templates for Figs. 1b, c, 2a, c are available through GenBank accession codes (see Methods).
Code availability
Code used to analyse sequencing datasets are available from the corresponding author upon request.
References
Gans, C. & Northcutt, R. G. Neural crest and the origin of vertebrates: a new head. Science 220, 268–273 (1983).
Glenn Northcutt, R. The new head hypothesis revisited. J. Exp. Zool. B Mol. Dev. Evol. 304, 274–297 (2005).
Simoes-Costa, M. & Bronner, M. E. Reprogramming of avian neural crest axial identity and cell fate. Science 352, 1570–1573 (2016).
Le Douarin, N. M. & Smith, J. Development of the peripheral nervous system from the neural crest. Annu. Rev. Cell Biol. 4, 375–404 (1988).
Le Douarin, N. The Neural Crest (Cambridge Univ. Press, 1982).
Sauka-Spengler, T., Meulemans, D., Jones, M. & Bronner-Fraser, M. Ancient evolutionary origin of the neural crest gene regulatory network. Dev. Cell 13, 405–420 (2007).
Nikitina, N., Sauka-Spengler, T. & Bronner-Fraser, M. Dissecting early regulatory relationships in the lamprey neural crest gene network. Proc. Natl Acad. Sci. USA 105, 20083–20088 (2008).
Green, S. A., Uy, B. R. & Bronner, M. E. Ancient evolutionary origin of vertebrate enteric neurons from trunk-derived neural crest. Nature 544, 88–91 (2017).
Häming, D. et al. Expression of sympathetic nervous system genes in Lamprey suggests their recruitment for specification of a new vertebrate feature. PLoS One 6, e26543 (2011).
Betancur, P., Bronner-Fraser, M. & Sauka-Spengler, T. Genomic code for Sox10 activation reveals a key regulatory enhancer for cranial neural crest. Proc. Natl Acad. Sci. USA 107, 3570–3575 (2010).
Haldin, C. E. & LaBonne, C. SoxE factors as multifunctional neural crest regulatory factors. Int. J. Biochem. Cell Biol. 42, 441–444 (2010).
McCauley, D. W. & Bronner-Fraser, M. Importance of SoxE in neural crest development and the evolution of the pharynx. Nature 441, 750–752 (2006).
Lee, E. M. et al. Functional constraints on SoxE proteins in neural crest development: the importance of differential expression for evolution of protein activity. Dev. Biol. 418, 166–178 (2016).
Green, S. A., Simoes-Costa, M. & Bronner, M. E. Evolution of vertebrates as viewed from the crest. Nature 520, 474–482 (2015).
Martik, M. L. & Bronner, M. E. Regulatory logic underlying diversification of the neural crest. Trends Genet. 33, 715–727 (2017).
Gillis, J. A., Alsema, E. C. & Criswell, K. E. Trunk neural crest origin of dermal denticles in a cartilaginous fish. Proc. Natl Acad. Sci. USA 114, 13200–13205 (2017).
Kent, W. J. BLAT—the BLAST-like alignment tool. Genome Res. 12, 656–664 (2002).
Sauka-Spengler, T. & Bronner-Fraser, M. Evolution of the neural crest viewed from a gene regulatory perspective. Genesis 46, 673–682 (2008).
Wada, H., Kobayashi, M. & Zhang, S. Ets identified as a trans-regulatory factor of amphioxus Hox2 by transgenic analysis using ascidian embryos. Dev. Biol. 285, 524–532 (2005).
Meulemans, D. & Bronner-Fraser, M. Amphioxus and lamprey AP-2 genes: implications for neural crest evolution and migration patterns. Development 129, 4953–4962 (2002).
Takahashi, T. & Holland, P. W. H. Amphioxus and ascidian Dmbx homeobox genes give clues to the vertebrate origins of midbrain development. Development 131, 3285–3294 (2004).
Yu, J.-K., Meulemans, D., McKeown, S. J. & Bronner-Fraser, M. Insights from the amphioxus genome on the origin of vertebrate neural crest. Genome Res. 18, 1127–1132 (2008).
Davidson, E. H. & Erwin, D. H. Gene regulatory networks and the evolution of animal body plans. Science 311, 796–800 (2006).
Parker, H. J., Bronner, M. E. & Krumlauf, R. A Hox regulatory network of hindbrain segmentation is conserved to the base of vertebrates. Nature 514, 490–493 (2014).
Parker, H. J., Bronner, M. E. & Krumlauf, R. The vertebrate Hox gene regulatory network for hindbrain segmentation: evolution and diversification. BioEssays 38, 526–538 (2016).
Nikitina, N., Bronner-Fraser, M. & Sauka-Spengler, T. Culturing lamprey embryos. Cold Spring Harbor Protoc. https://doi.org/10.1101/pdb.prot5122 (2009).
Di Tommaso, P. et al. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 39, W13–W17 (2011).
Armougom, F. et al. Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Res. 34, W604–W608 (2006).
O’Sullivan, O., Suhre, K., Abergel, C., Higgins, D. G. & Notredame, C. 3DCoffee: combining protein sequences and structures within multiple sequence alignments. J. Mol. Biol. 340, 385–395 (2004).
Poirot, O., Suhre, K., Abergel, C., O’Toole, E. & Notredame, C. 3DCoffee@igs: a web server for combining sequences and structures into a multiple sequence alignment. Nucleic Acids Res. 32, W37–W40 (2004).
Notredame, C., Higgins, D. G. & Heringa, J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217 (2000).
Longabaugh, W. J. R., Davidson, E. H. & Bolouri, H. Computational representation of developmental genetic regulatory networks. Dev. Biol. 283, 1–16 (2005).
Simões-Costa, M. S., McKeown, S. J., Tan-Cabugao, J., Sauka-Spengler, T. & Bronner, M. E. Dynamic and differential regulation of stem cell factor FoxD3 in the neural crest is encrypted in the genome. PLoS Genet. 8, e1003142 (2012).
Hamburger, V. & Hamilton, H. L. A series of normal stages in the development of the chick embryo. J. Morphol. 88, 49–92 (1951).
Smith, J. J. et al. The sea lamprey germline genome provides insights into programmed genome rearrangement and vertebrate evolution. Nat. Genet. 50, 270–277 (2018).
Karolchik, D. et al. The UCSC Genome Browser Database. Nucleic Acids Res. 31, 51–54 (2003).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Damas, H. Recherches sur le développement de Lampetra fluviatilis L. Contribution à l′étude de la céphalogenèse des vertébrés. Arch. Biol. (Liege) 1–284 (1944).
Modrell, M. S. et al. A fate-map for cranial sensory ganglia in the sea lamprey. Dev. Biol. 385, 405–416 (2014).
Acknowledgements
We thank J. Tan-Cabugao and E. Grossman for technical assistance; D. Mayorga and R. Fraser for help with fish husbandry; B. Martik for illustrating the adult animals for our expression matrices; the Caltech Millard and Muriel Jacobs Genetics and Genomics Laboratory and in particular I. Antoshechkin for sequencing of our RNA-seq libraries; and R. Diamond, J. Tijerina, D. Perez, and P. Cannon of the The Caltech Flow Cytometry Cell Sorting Facility for cell sorting assistance. This work is supported by NIH grants R01NS086907, R01DE024157, and R35NS111564 to M.E.B. M.L.M. is supported by a Helen Hay Whitney Foundation postdoctoral fellowship. S.G. is supported by a graduate fellowship from the American Heart Association (18PRE34050063).
Author information
Authors and Affiliations
Contributions
Project and analysis conception were designed by M.L.M., M.S.-C., and M.E.B. Writing and interpretation were performed by M.L.M., S.G., B.R.U., J.A.G., S.A.G., M.S.-C., and M.E.B. Lamprey orthologue cloning and all in situ hybridization, imaging, and analysis were performed by M.L.M. Bioinformatics and chicken RNA-seq were performed by S.G. Phylogenetic analysis and lamprey embryo acquisition were performed by S.A.G. Cloning of skate orthologues and skate embryo acquisition were performed by J.A.G. Lamprey embryo dissections and library preparations were performed by B.R.U. and M.S.C.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Peer review information Nature thanks Robert Cerny and Jeremiah Smith for their contribution to the peer review of this work.
Extended data figures and tables
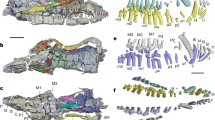
Extended Data Fig. 1 Heterochronic shifts of cranial specific gene regulatory nodes from later neural crest derivatives to an early specification program throughout gnathostome evolution.
a–d, Expression of lamprey orthologues of amniote cranial NC-specific genes at T21 (cranial) and T23 (trunk) in cross-section. e, Pharyngeal NC derivative expression in T26 P. marinus frontal section (based on ref. 40). f–l, Cranial circuit orthologues are expressed in pharyngeal arch derivatives, with the exception of Brn3, which is expressed in the NC-derived cranial sensory ganglia41 in lamprey frontal sections. m, Gene expression matrix summarizing the heterochronic shift of cranial crest specific circuit nodes. nc, neural crest; nt, neural tube; n, notochord. Scale bars, 100 µm. Cryosections of in situ hybridizations were reproducible on n ≥ 5 embryos per time point for n ≥ 2 experiments.
Extended Data Fig. 2 P. marinus Dmbx is homologous to gnathostome Dmbx genes.
a, Truncated alignment of Dmbx protein sequences. An alignment of full-length Dmbx protein sequences was assembled using TCoffee and contiguous regions tagged by the program as poorly or moderately well-aligned were removed, leaving 218 well-aligned residues. b, Bayesian consensus phylogenetic tree, with posterior probabilities shown at corresponding nodes.
Extended Data Fig. 3 Expression of cranial circuit genes in the neural crest of the little skate.
a, Schematic of a stage 18 L. erinacea embryo with the NC illustrated as blue (cranial) and red (trunk). b–f, Cross-sections as depicted in a. Scale bar, 50 µm. Cryosections of in situ hybridizations were reproducible on n ≥ 2 embryos for n ≥ 2 experiments.
Extended Data Fig. 4 Pharyngeal neural crest derivative expression of cranial circuit orthologues in stage 25 L. erinacea embryos.
a, Schematic of head of L. erinacea embryo. Dashed box represents the region of the head for each embryo shown in b–i (left); purple dashed line depicts the location of the frontal section for b–i (right). b–f, Right, pharyngeal NC derivative expression of cranial circuit orthologues. g–i, Right, Dmbx1, Lhx5, and Brn3c are absent in pharyngeal arch derivatives at stage 25. b–i, Scale bars, 500 µm (left); 100 µm (right). In situ hybridizations were reproducible on n ≥ 2 embryos.
Extended Data Fig. 5 Expression of cranial circuit genes in the NC of the zebrafish.
a, Schematic of a 14ss D. rerio embryo with the NC illustrated as blue (cranial) and red (trunk). b–h, Cross-sections as shown in a. Scale bars, 50 µm. In situ hybridizations were reproducible on n ≥ 10 embryos.
Extended Data Fig. 6 Expression of cranial circuit orthologues in pharyngeal NC derivatives of D. rerio embryos at 3 days post fertilization.
a, Schematic of head of zebrafish embryo at 3 days post fertilization (dpf). Purple dashed line depicts the location of the frontal sections for the insets in e–h. e–h, Expression of cranial circuit orthologues in pharyngeal arches. b–d, Dmbx1, Lhx5, and Brn3c are absent from pharyngeal arch derivatives at 3 dpf. Scale bar, 150 µm. In situ hybridizations were reproducible on n ≥ 10 whole-mount and cryosectioned embryos.
Supplementary information
Supplementary Table 1
Alignment percentage scores for lamprey/chicken alignments
Rights and permissions
About this article
Cite this article
Martik, M.L., Gandhi, S., Uy, B.R. et al. Evolution of the new head by gradual acquisition of neural crest regulatory circuits. Nature 574, 675–678 (2019). https://doi.org/10.1038/s41586-019-1691-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1691-4
This article is cited by
-
The hagfish genome and the evolution of vertebrates
Nature (2024)
-
Sea lamprey enlightens the origin of the coupling of retinoic acid signaling to vertebrate hindbrain segmentation
Nature Communications (2024)
-
Ascidian embryonic cells with properties of neural-crest cells and neuromesodermal progenitors of vertebrates
Nature Ecology & Evolution (2024)
-
Shaping faces: genetic and epigenetic control of craniofacial morphogenesis
Nature Reviews Genetics (2023)
-
Riding the crest to get a head: neural crest evolution in vertebrates
Nature Reviews Neuroscience (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.