Abstract
The large-scale genetic profiling of tumours can identify potentially actionable molecular variants for which approved anticancer drugs are available1,2,3. However, when patients with such variants are treated with drugs outside of their approved label, successes and failures of targeted therapy are not systematically collected or shared. We therefore initiated the Drug Rediscovery protocol, an adaptive, precision-oncology trial that aims to identify signals of activity in cohorts of patients, with defined tumour types and molecular variants, who are being treated with anticancer drugs outside of their approved label. To be eligible for the trial, patients have to have exhausted or declined standard therapies, and have malignancies with potentially actionable variants for which no approved anticancer drugs are available. Here we show an overall rate of clinical benefit—defined as complete or partial response, or as stable disease beyond 16 weeks—of 34% in 215 treated patients, comprising 136 patients who received targeted therapies and 79 patients who received immunotherapy. The overall median duration of clinical benefit was 9 months (95% confidence interval of 8–11 months), including 26 patients who were experiencing ongoing clinical benefit at data cut-off. The potential of the Drug Rediscovery protocol is illustrated by the identification of a successful cohort of patients with microsatellite instable tumours who received nivolumab (clinical benefit rate of 63%), and a cohort of patients with colorectal cancer with relatively low mutational load who experienced only limited clinical benefit from immunotherapy. The Drug Rediscovery protocol facilitates the defined use of approved drugs beyond their labels in rare subgroups of cancer, identifies early signals of activity in these subgroups, accelerates the clinical translation of new insights into the use of anticancer drugs outside of their approved label, and creates a publicly available repository of knowledge for future decision-making.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
All data described in this study are freely available for academic use, and can be obtained through the Netherlands Cancer Institute and the HMF through standardized procedures and request forms. These can be found at https://www.hartwigmedicalfoundation.nl/en. In brief, a data request can be initiated by filling out the standard form, which requires the applicant to provide the intended use of the requested data. Next, an independent data-access board will evaluate whether the intended use of the data is compatible with the consent given by the patients, and whether there would be any applicable legal or ethical constraints. Upon formal approval by the data-access board, a standard license agreement (which does not have any restrictions regarding intellectual property resulting from the data analysis) needs to be signed by an official representation of the organization before access to the data is granted. All WGS data, including raw data files, will be made available through a dedicated download portal with two-factor authentication, via the HMF. Clinical, outcome and safety data can be obtained at a per-patient level for all patients reported in the study, by emailing the Institutional Review Board of the Netherlands Cancer Institute (IRB@nki.nl). Both the WGS and clinical datasets have unique identifiers for individual patients, which allows a combined analysis. For any other data, correspondence should be addressed to e.voest@nki.nl and requests for materials to DRUP@nki.nl.
References
Hainsworth, J. D. et al. Targeted therapy for advanced solid tumors on the basis of molecular profiles: results from MyPathway, an open-label, phase IIa multiple basket study. J. Clin. Oncol. 36, 536–542 (2018).
Hyman, D. M. et al. HER kinase inhibition in patients with HER2- and HER3-mutant cancers. Nature 554, 189–194 (2018).
Hyman, D. M. et al. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N. Engl. J. Med. 373, 726–736 (2015).
Colwell, J. NCI-MATCH trial draws strong interest. Cancer Discov. 6, 334 (2016).
Massard, C. et al. High-throughput genomics and clinical outcome in hard-to-treat advanced cancers: results of the MOSCATO 01 trial. Cancer Discov. 7, 586–595 (2017).
Meric-Bernstam, F. et al. Feasibility of large-scale genomic testing to facilitate enrollment onto genomically matched clinical trials. J. Clin. Oncol. 33, 2753–2762 (2015).
Stockley, T. L. et al. Molecular profiling of advanced solid tumors and patient outcomes with genotype-matched clinical trials: the Princess Margaret IMPACT/COMPACT trial. Genome Med. 8, 109 (2016).
Hyman, D. M., Taylor, B. S. & Baselga, J. Implementing genome-driven oncology. Cell 168, 584–599 (2017).
Le Tourneau, C. et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 16, 1324–1334 (2015).
Prasad, V. Perspective: the precision-oncology illusion. Nature 537, S63 (2016).
Tannock, I. F. & Hickman, J. A. Limits to personalized cancer medicine. N. Engl. J. Med. 375, 1289–1294 (2016).
Sleijfer, S., Bogaerts, J. & Siu, L. L. Designing transformative clinical trials in the cancer genome era. J. Clin. Oncol. 31, 1834–1841 (2013).
Ellis, L. M. et al. American Society of Clinical Oncology perspective: raising the bar for clinical trials by defining clinically meaningful outcomes. J. Clin. Oncol. 32, 1277–1280 (2014).
Bins, S. et al. Implementation of a multicenter biobanking collaboration for next-generation sequencing-based biomarker discovery based on fresh frozen pretreatment tumor tissue biopsies. Oncologist 22, 33–40 (2016).
Simon, R. Optimal two-stage designs for phase II clinical trials. Control. Clin. Trials 10, 1–10 (1989).
Jung, S. H., Lee, T., Kim, K. & George, S. L. Admissible two-stage designs for phase II cancer clinical trials. Stat. Med. 23, 561–569 (2004).
Moreno Garcia, V. et al. Dose–response relationship in phase I clinical trials: a European Drug Development Network (EDDN) collaboration study. Clin. Cancer Res. 20, 5663–5671 (2014).
Le, D. T. et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 372, 2509–2520 (2015). 10
Overman, M. J. et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 18, 1182–1191 (2017).
van Waalwijk van Doorn-Khosrovani, S. B. et al. Personalised reimbursement: a risk-sharing model for biomarker-driven treatment of rare subgroups of cancer patients. Ann. Oncol. 30, 663–665 (2019).
Priestley, P. et al. Pan-cancer whole genome analyses of metastatic solid tumors. Preprint at https://www.biorxiv.org/content/10.1101/415133v4 (2018).
Griffith, M. et al. CIViC is a community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer. Nat. Genet. 49, 170–174 (2017).
Chakravarty, D. et al. OncoKB: a precision oncology knowledge base. JCO Precis. Oncol. https://doi.org/10.1200/PO.17.00011 (2017).
Tamborero, D. et al. Cancer Genome Interpreter annotates the biological and clinical relevance of tumor alterations. Genome Med. 10, 25 (2018).
Mateo, J. et al. A framework to rank genomic alterations as targets for cancer precision medicine: the ESMO scale for clinical actionability of molecular targets (ESCAT). Ann. Oncol. 29, 1895–1902 (2018).
Sleijfer, S. & Wagner, A. J. The challenge of choosing appropriate end points in single-arm phase II studies of rare diseases. J. Clin. Oncol. 30, 896–898 (2012).
Ray, T. CMS-proposed coverage of NGS cancer tests could lead to off-label scripts, oncologists worry, https://www.genomeweb.com/molecular-diagnostics/cms-proposed-coverage-ngs-cancer-tests-could-lead-label-scripts-oncologists#.WsJTUOkUk5k (2018).
Meric-Bernstam, F. et al. A decision support framework for genomically informed investigational cancer therapy. J. Natl Cancer Inst. 107, djv098 (2015).
Cheson, B. D. et al. Recommendations for initial evaluation, staging, and response assessment of Hodgkin and non-Hodgkin lymphoma: the Lugano classification. J. Clin. Oncol. 32, 3059–3067 (2014).
Rajkumar, S. V. et al. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 15, e538–e548 (2014).
Rustin, G. J. et al. Definitions for response and progression in ovarian cancer clinical trials incorporating RECIST 1.1 and CA 125 agreed by the Gynecological Cancer Intergroup (GCIG). Int. J. Gynecol. Cancer 21, 419–423 (2011).
Therasse, P. et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J. Natl. Cancer Inst. 92, 205–216 (2000).
Wen, P. Y. et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J. Clin. Oncol. 28, 1963–1972 (2010).
Huang, M. N. et al. MSIseq: software for assessing microsatellite instability from catalogs of somatic mutations. Sci. Rep. 5, 13321 (2015).
Acknowledgements
We thank the Barcode for Life Foundation and the Dutch Cancer Society for their financial support; Amgen, Astra Zeneca, Bayer, Boehringer Ingelheim, Bristol-Myers Squibb, Eisai, Merck Sharp and Dohme, Novartis, Pfizer and Roche for their in-kind and financial support; the HMF for their in-kind support by performing sequencing and biomarker analyses on baseline biopsies; the Center for Personalized Cancer Treatment Multidisciplinary Expert Board for supporting the central case-review process; the Independent Data Monitoring Committee for their advice on cohort decisions and the monitoring of preliminary safety data; the Netherlands Cancer Institute’s Biobank Facility, Scientific Department and Pharmacy for their facilitating services; A. P. Hamberg, L. V. Beerepoot and J. M. Meerum-Terwogt for their contributions to trial recruitment; and all participating hospitals for supporting and facilitating the conduct of the DRUP trial.
Author information
Authors and Affiliations
Contributions
All authors contributed extensively to the work presented in this paper. H.M.W.V., H.G. and E.E.V. initiated and led the trial as principal investigators. R.L.S. and S.S. contributed to its design and initiation. D.L.v.d.V., L.R.H., H.v.d.W. and J.M.v.B.H. coordinated the trial. D.L.v.d.V. analysed the data and wrote the manuscript. L.R.H., H.v.d.W. and J.M.v.B.H. assisted with the data analysis, and substantively revised the manuscript. C.M.L.v.H., D.J.A.d.G., L.A.D., A.H., M.J.A.d.J., M.C., E.F.S., A.J.d.L., N.M., M.L. and E.K. contributed to patient enrolment and clinical data collection. P.R., W.W.J.d.L. and P.M.N. helped with the interpretation of tumour profiles by performing variant calling and pathogenicity assessments. A.D.R.H. and B.N. coordinated the distribution of study drugs. P.R. and E.C. performed sequencing of tumour biopsies, generated sequencing reports and conducted biomarker analyses. E.v.W. contributed to data extractions, and statistical design and analyses. All authors discussed the results and implications, and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
E.E.V. is legally responsible for all contracts with pharmaceutical companies at the Netherlands Cancer Institute. H.M.W.V. and H.G. have, through the DRUP and other studies, received support from pharmaceutical companies that are participating in the DRUP. R.L.S serves as principal investigator for the TAPUR trial, in support of which the American Society of Clinical Oncology receives financial support from Astra-Zeneca, Bayer, Bristol-Myers Squibb, Genentech, Lilly, Merck and Pfizer. C.M.L.v.H. has received funding from AstraZeneca, BMS, Ipsen, Merck, MSD and Novartis. A.J.d.L. has served as an advisor for AstraZeneca, BMS, Boehringer, Pfizer, Lilly and MSD, and has received research grants from AstraZeneca, BMS and MSD. E.K. has served as advisor for Amgen, BMS, Eisai, Genzyme-Sanofi, MSD, Novartis and Roche. The other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Peer review information Nature thanks David Hyman, Christophe Le Tourneau and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
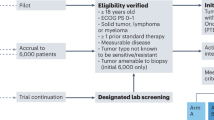
Extended Data Fig. 1 Study flowchart.
Patients may be identified via regular diagnostics or by WGS performed within the context of a CPCT sequencing study. Adult patients with advanced cancers and without standard-treatment options (but with a known potentially actionable variant in their molecular tumour profile) can be submitted for review. The central review is done by two or more reviewers independently, supported by the CPCT Molecular Tumour Board, and includes review of (i) the medical history of the patient, (ii) tumour-profiling test results, (iii) available literature and (iv) potential drug-access alternatives. Patients who are eligible for standard treatments are referred back to their treating physician. Genomic variants of unknown significance (VUS) that are not likely to be actionable are not considered acceptable drug targets. Negative trials are not repeated, nor are positive or ongoing phase II or III trials, unless drug access is not (or is not yet) facilitated. Drug access via other trials or access programmes is preferred, if available. Input for stages (iii) and (iv) of the review is derived from PubMed (https://www.ncbi.nlm.nih.gov/pubmed/), ClinicalTrials.gov and weekly automatic updates on publications that mention any drug in the study in their titles and/or abstracts. If the general selection criteria are met and the appropriate study treatment is available, the patient can be informed, screened and enrolled (if all drug-specific selection criteria are also met). Once a fresh baseline tumour biopsy is obtained, study treatment can be initiated. Patients are treated and followed according to the labelled indication for each drug. Response is evaluated once every two months. Patients can continue study treatment as long as clinical benefit is observed. Patients who discontinue study treatment can be resubmitted if their molecular tumour profile (as revealed by the baseline biopsy) contains additional actionable variants. CR, complete response, PR, partial response.
Extended Data Fig. 2 Case submissions and reasons for non-accrual.
Overview of the first 642 case submissions (submitted between 1 September 2016 and 1 September 2018), as well as the reasons for not being enrolled in the study. Values are displayed as a percentage relative to these 642 case submissions, and as an absolute number per category. Cases that were erroneously submitted (owing to incomplete understanding of the study protocol and/or retraction of the submission by the treating physician) are not included in this overview (n = 58 cases).
Extended Data Fig. 3 Baseline biopsies for biomarker analyses.
Overview and success rate of WGS on pre-treatment tumour biopsies. The bottom panel displays the number of patients for whom WGS succeeded, and indicates whether the initial variant (on the basis of which the patient started the study treatment) was also present in the fresh baseline biopsy. Values are displayed as absolute numbers and percentages, relative to the 131 successfully sequenced biopsies. CPCT-02, the national WGS programme of the CPCT; HML, high mutational load (defined as ≥140 somatic missense variants across the tumour genome overall); WGS-MSI, microsatellite instability suspected on the basis of WGS results; ampl, amplification; mut, mutation; wt, wild type.
Supplementary information
Supplementary Table
Supplementary Table 1: Tumour profiles and Whole Genome Sequencing. This table summarizes the molecular tumour profile of all enrolled patients for whom WGS data were available and the potentially relevant additional WGS findings per patient.
Rights and permissions
About this article
Cite this article
van der Velden, D.L., Hoes, L.R., van der Wijngaart, H. et al. The Drug Rediscovery protocol facilitates the expanded use of existing anticancer drugs. Nature 574, 127–131 (2019). https://doi.org/10.1038/s41586-019-1600-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1600-x
This article is cited by
-
Multiregion WES of metastatic pancreatic neuroendocrine tumors revealed heterogeneity in genomic alterations, immune microenvironment and evolutionary patterns
Cell Communication and Signaling (2024)
-
A federated learning system for precision oncology in Europe: DigiONE
Nature Medicine (2024)
-
Comprehensive genomic profiling on metastatic Melanoma: results from a network screening from 7 Italian Cancer Centres
Journal of Translational Medicine (2024)
-
Optimized whole-genome sequencing workflow for tumor diagnostics in routine pathology practice
Nature Protocols (2024)
-
Genome-matched treatments and patient outcomes in the Maine Cancer Genomics Initiative (MCGI)
npj Precision Oncology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.