Abstract
Many wastewater treatment plants around the world suffer from the operational problem of foaming. This is characterized by a persistent stable foam that forms on the aeration basin, which reduces effluent quality. The foam is often stabilized by a highly hydrophobic group of Actinobacteria known as the Mycolata1. Gordonia amarae is one of the most frequently reported foaming members1. With no currently reliable method for treating foams, phage biocontrol has been suggested as an attractive treatment strategy2. Phages isolated from related foaming bacteria can destabilize foams at the laboratory scale3,4; however, no phage has been isolated that lyses G. amarae. Here, we assemble the complete genomes of G. amarae and a previously undescribed species, Gordonia pseudoamarae, to examine mechanisms that encourage stable foam production. We show that both of these species are recalcitrant to phage infection via a number of antiviral mechanisms including restriction, CRISPR–Cas and bacteriophage exclusion. Instead, we isolate and cocultivate an environmental ultrasmall epiparasitic bacterium from the phylum Saccharibacteria that lyses G. amarae and G. pseudoamarae and several other Mycolata commonly associated with wastewater foams. The application of this parasitic bacterium, ‘Candidatus Mycosynbacter amalyticus’, may represent a promising strategy for the biocontrol of bacteria responsible for stabilizing wastewater foams.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The bacterial genomes assembled in this study are available on the National Institutes of Health GenBank under the following accession numbers: G. amarae CON44T, CP045810; G. amarae BEN368, CP045808; G. amarae BEN372 CP045805; G. amarae BEN374, CP045804; G. pseudoamarae CON9T, CP045809; G. pseudoamarae BEN371, CP045806 and CP045807; and M. amalyticus JR1, CP045921. Raw sequence data are available on the NCBI under BioProject accession no. PRJNA587209. Bacterial strains are available from the ATCC, DSMZ and JCM culture collections or from the authors upon request. Source data are provided with this paper.
References
Seviour, R. J. & Nielsen, P. H. Microbial Ecology of Activated Sludge (IWA Publishing, 2010).
Batinovic, S. et al. Bacteriophages in natural and artificial environments. Pathogens 8, 100 (2019).
Petrovski, S., Seviour, R. J. & Tillett, D. Prevention of Gordonia and Nocardia stabilized foam formation by using bacteriophage GTE7. Appl. Environ. Microbiol. 77, 7864–7867 (2011).
Petrovski, S., Seviour, R. J. & Tillett, D. Characterization of the genome of the polyvalent lytic bacteriophage GTE2, which has potential for biocontrol of Gordonia-, Rhodococcus-, and Nocardia-stabilized foams in activated sludge plants. Appl. Environ. Microbiol. 77, 3923–3929 (2011).
Nielsen, P. H., Kragelund, C., Seviour, R. J. & Nielsen, J. L. Identity and ecophysiology of filamentous bacteria in activated sludge. FEMS Microbiol. Rev. 33, 969–998 (2009).
De los Reyes, F. in Microbial Ecology of Activated Sludge (eds Seviour, R. J. & Nielsen, P. H.) 215–258 (IWA Publishing, 2010).
Wanner, J., Kragelund, C. & Nielsen, P. H. in Microbial Ecology of Activated Sludge (eds Seviour, R. J. & Nielsen, P. H.) 191–214 (IWA publishing, 2010).
Soddell, J. & Seviour, R. J. Microbiology of foaming in activated sludge plants. J. Appl. Bacteriol. 69, 145–176 (1990).
Tandoi, V., Rossetti, S. & Wanner, J. Activated Sludge Separation Problems: Theory, Control Measures, Practical Experiences (IWA Publishing, 2017).
Kragelund, C. et al. Ecophysiology of mycolic acid-containing Actinobacteria (Mycolata) in activated sludge foams. FEMS Microbiol. Ecol. 61, 174–184 (2007).
Lechevalier, M. P. & Lechevalier, H. A. Nocardia amarae sp. nov., an actinomycete common in foaming activated sludge. Int. J. Syst. Bacteriol. 24, 278–288 (1974).
de los Reyes, F. L. III, Rothauszky, D. & Raskin, L. Microbial community structures in foaming and nonfoaming full-scale wastewater treatment plants. Water Environ. Res. 74, 437–449 (2002).
Winkler, M. K., Kröber, E., Mohn, W. W., Koch, F. & Frigon, D. Comparison of microbial populations and foaming dynamics in conventional versus membrane enhanced biological phosphorous removal systems. Water Environ. J. 30, 102–112 (2016).
Pal, P., Khairnar, K. & Paunikar, W. N. Causes and remedies for filamentous foaming in activated sludge treatment plant. Glob. Nest J. 16, 762–772 (2014).
Lemmer, H. & Kroppenstedt, R. M. Chemotaxonomy and physiology of some actinomycetes isolated from scumming activated-sludge. Syst. Appl. Microbiol. 5, 124–135 (1984).
Auch, A. F., von Jan, M., Klenk, H.-P. & Göker, M. Digital DNA–DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genom. Sci. 2, 117–134 (2010).
Goris, J. et al. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91 (2007).
Konstantinidis, K. T. & Tiedje, J. M. Towards a genome-based taxonomy for prokaryotes. J. Bacteriol. 187, 6258–6264 (2005).
Chaumeil, P.-A., Mussig, A. J., Hugenholtz, P. & Parks, D. H. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36, 1925–1927 (2020).
Petrovski, S., Tillett, D. & Seviour, R. J. Genome sequences and characterization of the related Gordonia phages GTE5 and GRU1 and their use as potential biocontrol agents. Appl. Environ. Microbiol. 78, 42–47 (2012).
Dyson, Z. A., Tucci, J., Seviour, R. J. & Petrovski, S. Lysis to kill: evaluation of the lytic abilities, and genomics of nine bacteriophages infective for Gordonia spp. and their potential use in activated sludge foam biocontrol. PLoS ONE 10, e0134512 (2015).
Russell, D. A. & Hatfull, G. F. PhagesDB: the actinobacteriophage database. Bioinformatics 33, 784–786 (2017).
Goldfarb, T. et al. BREX is a novel phage resistance system widespread in microbial genomes. EMBO J. 34, 169–183 (2015).
He, X. et al. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc. Natl Acad. Sci. USA 112, 244–249 (2015).
Cross, K. L. et al. Targeted isolation and cultivation of uncultivated bacteria by reverse genomics. Nat. Biotechnol. 37, 1314–1321 (2019).
Collins, A. J., Murugkar, P. P. & Dewhirst, F. E. Complete genome sequence of strain AC001, a novel cultured member of the human oral microbiome from the candidate phylum Saccharibacteria (TM7). Microbiol. Resour. Announc. 8, e01158-19 (2019).
Brown, C. T. et al. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523, 208–211 (2015).
Bor, B. et al. Rapid evolution of decreased host susceptibility drives a stable relationship between ultrasmall parasite TM7x and its bacterial host. Proc. Natl Acad. Sci. USA 115, 12277–12282 (2018).
Albertsen, M. et al. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat. Biotechnol. 31, 533–538 (2013).
Starr, E. P. et al. Stable isotope informed genome-resolved metagenomics reveals that Saccharibacteria utilize microbially-processed plant-derived carbon. Microbiome 6, 122 (2018).
Kantor, R. S. et al. Small genomes and sparse metabolisms of sediment-associated bacteria from four candidate phyla. mBio 4, e00708-13 (2013).
McLean, J. S. et al. Acquisition and adaptation of ultra-small parasitic reduced genome bacteria to mammalian hosts. Cell Rep. 32, 107939 (2020).
Mattick, J. S. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56, 289–314 (2002).
Bernheim, A. & Sorek, R. The pan-immune system of bacteria: antiviral defence as a community resource. Nat. Rev. Microbiol. 18, 113–119 (2020).
Borneman, J. & Triplett, E. W. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl. Environ. Microbiol. 63, 2647–2653 (1997).
Zhang, L. et al. Bacterial and archaeal communities in the deep-sea sediments of inactive hydrothermal vents in the Southwest India Ridge. Sci. Rep. 6, 25982 (2016).
Dewhirst, F. E. et al. The human oral microbiome. J. Bacteriol. 192, 5002–5017 (2010).
Bond, P. L., Hugenholtz, P., Keller, J. & Blackall, L. L. Bacterial community structures of phosphate-removing and non-phosphate-removing activated sludges from sequencing batch reactors. Appl. Environ. Microbiol. 61, 1910–1916 (1995).
Castelle, C. J. et al. Biosynthetic capacity, metabolic variety and unusual biology in the CPR and DPANN radiations. Nat. Rev. Microbiol. 16, 629–645 (2018).
Hahn, H. P. The type-4 pilus is the major virulence-associated adhesin of Pseudomonas aeruginosa—a review. Gene 192, 99–108 (1997).
Mazariego-Espinosa, K., Cruz, A., Ledesma, M. A., Ochoa, S. A. & Xicohtencatl-Cortes, J. Longus, a type IV pilus of enterotoxigenic Escherichia coli, is involved in adherence to intestinal epithelial cells. J. Bacteriol. 192, 2791–2800 (2010).
Brown, D. R., Helaine, S., Carbonnelle, E. & Pelicic, V. Systematic functional analysis reveals that a set of seven genes is involved in fine-tuning of the multiple functions mediated by type IV pili in Neisseria meningitidis. Infect. Immun. 78, 3053–3063 (2010).
Rodgers, K., Arvidson, C. G. & Melville, S. Expression of a Clostridium perfringens type IV pilin by Neisseria gonorrhoeae mediates adherence to muscle cells. Infect. Immun. 79, 3096–3105 (2011).
Evans, K. J., Lambert, C. & Sockett, R. E. Predation by Bdellovibrio bacteriovorus HD100 requires type IV pili. J. Bacteriol. 189, 4850–4859 (2007).
Mahmoud, K. K. & Koval, S. F. Characterization of type IV pili in the life cycle of the predator bacterium Bdellovibrio. Microbiology (Reading) 156, 1040–1051 (2010).
Chanyi, R. M. & Koval, S. F. Role of type IV pili in predation by Bdellovibrio bacteriovorus. PLoS ONE 9, e113404 (2014).
Wong, W. Y. et al. Structure-function analysis of the adherence-binding domain on the pilin of Pseudomonas aeruginosa strains PAK and KB7. Biochemistry 34, 12963–12972 (1995).
Utter, D. R., He, X., Cavanaugh, C. M., McLean, J. S. & Bor, B. The saccharibacterium TM7x elicits differential responses across its host range. ISME J. 14, 3054–3067 (2020).
Skerman, V. A new type of micromanipulator and microforge. J. Gen. Microbiol. 54, 287–297 (1968).
Petrovski, S., Seviour, R. J. & Tillett, D. Genome sequence and characterization of the Tsukamurella bacteriophage TPA2. Appl. Environ. Microbiol. 77, 1389–1398 (2011).
Santos, M. A. An improved method for the small scale preparation of bacteriophage DNA based on phage precipitation by zinc chloride. Nucleic Acids Res. 19, 5442 (1991).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011).
Wick, R. R., Judd, L. M., Gorrie, C. L. & Holt, K. E. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, e1005595 (2017).
Stecher, G., Tamura, K. & Kumar, S. Molecular evolutionary genetics analysis (MEGA) for macOS. Mol. Biol. Evol. 37, 1237–1239 (2020).
Saitou, N. & Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Felsenstein, J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791 (1985).
Tamura, K. & Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526 (1993).
Fitch, W. M. Toward defining the course of evolution: minimum change for a specific tree topology. Syst. Zool. 20, 406–416 (1971).
Richter, M., Rosselló-Móra, R., Oliver Glöckner, F. & Peplies, J. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32, 929–931 (2016).
Rodriguez-R, L. M. & Konstantinidis, K. T. The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. Preprint at PeerJ Preprints https://doi.org/10.7287/peerj.preprints.1900v1 (2016).
Meier-Kolthoff, J. P. & Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 10, 2182 (2019).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2019).
Galili, T., O’Callaghan, A., Sidi, J. & Sievert, C. heatmaply: an R package for creating interactive cluster heatmaps for online publishing. Bioinformatics 34, 1600–1602 (2018).
Darling, A. E., Mau, B. & Perna, N. T. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 5, e11147 (2010).
Guy, L., Roat Kultima, J. & Andersson, S. genoPlotR: comparative gene and genome visualization in R. Bioinformatics 26, 2334–2335 (2010).
Page, A. J. et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693 (2015).
Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).
Huerta-Cepas, J. et al. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 34, 2115–2122 (2017).
Huerta-Cepas, J. et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314 (2019).
Tatusova, T. et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624 (2016).
Yu, Y., Ouyang, Y. & Yao, W. shinyCircos: an R/Shiny application for interactive creation of Circos plot. Bioinformatics 34, 1229–1231 (2018).
Zimmermann, L. et al. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 430, 2237–2243 (2018).
Roberts, R. J., Vincze, T., Posfai, J. & Macelis, D. REBASE—a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 43, D298–D299 (2015).
Couvin, D. et al. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 46, W246–W251 (2018).
Vallenet, D. et al. MicroScope: an integrated platform for the annotation and exploration of microbial gene functions through genomic, pangenomic and metabolic comparative analysis. Nucleic Acids Res. 48, D579–D589 (2020).
Imam, S., Chen, Z., Roos, D. S. & Pohlschröder, M. Identification of surprisingly diverse type IV pili, across a broad range of gram-positive bacteria. PLoS ONE 6, e28919 (2011).
Rawlings, N. D., Waller, M., Barrett, A. J. & Bateman, A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 42, D503–D509 (2014).
Janecek, M. & Kral, R. Modern Electron Microscopy in Physical and Life Sciences (InTech, 2016).
Miłobędzka, A. & Muszyński, A. Population dynamics of filamentous bacteria identified in Polish full-scale wastewater treatment plants with nutrients removal. Water Sci. Technol. 71, 675–684 (2015).
Jo, S. J., Kwon, H., Jeong, S.-Y., Lee, C.-H. & Kim, T. G. Comparison of microbial communities of activated sludge and membrane biofilm in 10 full-scale membrane bioreactors. Water Res. 101, 214–225 (2016).
Stokholm-Bjerregaard, M. et al. A critical assessment of the microorganisms proposed to be important to enhanced biological phosphorus removal in full-scale wastewater treatment systems. Front. Microbiol. 8, 718 (2017).
Henriet, O., Meunier, C., Henry, P. & Mahillon, J. Filamentous bulking caused by Thiothrix species is efficiently controlled in full-scale wastewater treatment plants by implementing a sludge densification strategy. Sci. Rep. 7, 1430 (2017).
Chen, Y.-L. et al. Estrogen degraders and estrogen degradation pathway identified in an activated sludge. Appl. Environ. Microbiol. 84, e00001-18 (2018).
Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019).
Callahan, B. J. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
McIlroy, S. J. et al. MiDAS: the field guide to the microbes of activated sludge. Database (Oxford) 2015, bav062 (2015).
Acknowledgements
We thank E. Johnston for helpful discussions and S. Knowler for assistance with R and QIIME 2. J.J.A.R. was supported by a La Trobe University postgraduate award. This work was funded by university funds provided to the department. The authors received no specific funding for this work.
Author information
Authors and Affiliations
Contributions
S.B. and S.P. conceived and designed the study. S.B. performed the WGS, assembly and analysis of G. amarae, G. pseudoamarae and M. amalyticus. J.J.A.R. isolated M. amalyticus and performed the coculture and host range experiments. J.R. performed the electron microscopy. S.B. wrote the manuscript in consultation with J.J.A.R., R.J.S. and S.P. All authors read and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Microbiology thanks Xuesong He, Per H. Nielsen, Mircea Podar and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
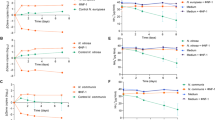
Extended Data Fig. 1 Whole genome comparative analyses leads to reclassification of two G. amarae strains as G. pseudoamarae.
a, BLASTn whole genome sequence comparison showing pairwise % coverage and identity between six strains sequenced in this study. b, Whole genome alignment of CON44T and CON9T. Horizontal colored blocks represent a genome and vertical lines represent homologous regions between different strains. Dark blue lines indicate regions of genome inversion. Scale bar indicate 1 Mb. c, Heatmap representing percentage average amino acid identity (AAI) analysis of the whole genomes of six Gordonia strains assembled in this study and six related Gordonia species genomes obtained from NCBI. d, Phylogenetic reconstruction based on the amino acid sequence of a concatenated 120 standard bacterial protein set from G. amarae CON44T, G. pseudoamarae CON9T and six related Gordonia species (Supplementary Table 8) generated using GTDB-Tk. The phylogenetic tree is inferred using the neighbor-joining method with a bootstrap test (1000 replicates). Phylogenetic trees were also reconstructed using the maximum parsimony and maximum likelihood methods with bootstrap tests (1000 replicates). Filled circles indicate the corresponding nodes were present in all three methods. Tsukamurella paurometabola DSM 20162T was used as an outgroup. Bar represents the number of amino acid substitutions per site.
Extended Data Fig. 2 Functional categorization of the core proteome of G. amarae and G. pseudoamarae.
a, b, Core protein analysis (>95% amino acid identity) performed on a, 4 G. amarae genomes (CON44T, BEN368, BEN372 and BEN374) and b, 2 G. pseudoamarae genomes (CON9T and BEN371) identified a core proteome of 4533 and 4299 proteins, respectively. COG analysis was used to sort these proteins into functional groups.
Extended Data Fig. 3 Identification of genes involved in the synthesis and export of mycolic acid.
Homology-based analysis using M. tuberculosis J37Rv as a model revealed orthologues of genes required for synthesis and export of mycolic acid in G. amarae and G. pseudoamarae species sequenced here, and seven additional Gordonia spp. Green shaded cells represent genes for which orthologues were identified. Full detail available in Supplementary Table 1.
Extended Data Fig. 4 Schematic of active CRISPR-Cas loci in G. amarae and G. pseudoamarae.
Schematic of the CRISPR-Cas locus in a, G. amarae CON44T (Type I-U) and b, G. pseudoamarae CON9T (Type I-E). Spacer region (red diamonds) is expanded below each locus showing alignment of CRISPR spacers from each strain. Sequence identity and scale is show. Repeat sequences are colored in grey. Spacers are color coded to demonstrate the same spacer present between strains. Yellow colored spacers represent unique spacers not found in other strains.
Extended Data Fig. 5 Genomic characterization and comparison of M. amalyticus.
a, Plot of the complete genome sequence of M. amalyticus JR1. From outer to inner: blue track represents sequencing coverage using a sliding window of 1,000 bp. Outer grey circle indicates maximum coverage (412-fold) and inner grey circle indicates average coverage (315-fold). The orange and green tracks indicate genes encoded in the forward and reverse strands, respectively. The light blue track indicates tRNA genes, and the red track shows the location of the single rRNA operon. Finally, the purple track represents GC content using a sliding window of 1,000 bp with the outer and inner grey circles indicating maximum (62%) and average (50.4%) GC content, respectively. Scale of genome length in Mb is shown inside b, BLASTn analysis of full-length M. amalyticus JR1 16 S rRNA gene showing % identity of top five closest hits. c, Phylogenetic reconstruction based on the amino acid sequence of a concatenated 120 bacterial standard protein set from M. amalyticus JR1 and 27 related Saccharibacteria (Supplementary Table 8) generated using GTDB-Tk. The phylogenetic tree is inferred using the neighbor-joining method with a bootstrap test (1000 replicates). Ca. Parcubacteria RAAC4-OD1 was used as an outgroup. Bar represents the number of amino acid substitutions per site. d, Heatmap based on percentage amino acid identity analysis of the genome sequence of M. amalyticus JR1 M. amalyticus and related Saccharibacteria as per the above panel. Text highlighted in green indicate Saccharibacteria that were identified in external environments (that is not from the human oral environment).
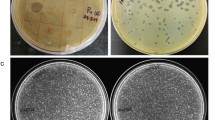
Extended Data Fig. 6 Host range of M. amalyticus.
a, Serial tenfold dilutions (106-103 PFU ml−1) of M. amalyticus JR1 were spotted onto a lawn of G. pseudoamarae CON9T grown on solid medium. Clearing of the CON9T lawn is observed in regions where M. amalyticus is spotted. Full clearing is observed at 1×107-106 PFU ml−1 whereas individual plaques are visible at lower concentrations (1×104 PFU ml−1). Image is representative of three biological replicates. b, Host range analysis of M. amalyticus JR1 on bacteria from the Mycolata group. Serial tenfold dilutions (108-105 PFU ml−1) were spotted onto bacterial lawn plates grown on solid medium. Species that exhibited lysis (full clearing at 1×107-106 ml−1 and individual plaques at 1×104-103 ml−1) were considered to be sensitive to M. amalyticus JR1.
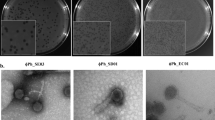
Extended Data Fig. 7 Additional examples of pili structures on M. amalyticus.
Black arrows indicate pili structures extending from cells. Scale bars, 500 nm. Images are representative of two independent experiments.
Extended Data Fig. 8 Growth curve of CON44T and M. amalyticus coculture.
Isolated M. amalyticus cocultured with CON44T (OD600 = 0.05) at a MOI of 0.1 over 66 hours (X axis) to determine the effect of M. amalyticus on host growth. A CON44T monoculture was used as a control. Data are presented as mean values (solid line) ± SEM (dotted lines) with individual data points represented by closed circles (n = 4 biologically independent experiments).
Extended Data Fig. 9 Description of T4SS genes in M. amalyticus.
a, Details of the T4SS genes identified in M. amalyticus with corresponding Gene ID. b, Number of predicted pilins identified in M. amalyticus, and 25 additional Ca. Saccharibacteria genomes (Supplementary Table 8) as identified by PilFind. Text highlighted in green indicate Saccharibacteria that were identified in external environments (that is not from the human oral environment).
Extended Data Fig. 10 Phylogenies of selected Actinobacteria hosts and Saccharibacteria genomes.
On the left is a phylogenetic tree reconstructed based on the 16 S rRNA gene sequence from 43 Actinobacteria species (Supplementary Table 8) that have been associated with Saccharibacteria studies using the neighbor-joining method with a bootstrap test (1000 replicates). Bar represents the number of nucleotide substitutions per site. On the right is a phylogenetic tree (from Extended Data Fig. 5c) reconstructed based on the amino acid sequence of a concatenated 120 standard bacterial protein set from 27 Saccharibacteria genomes (Supplementary Table 8) generated by GTDB-Tk using the neighbor-joining method with a bootstrap test (1000 replicates). Bar represents the number of amino acid substitutions per site. Matching coloured stars and circles indicate the host is susceptible to lysis by that Saccharibacteria. Red text indicates human oral strains cocultivated in the laboratory while green text indicates non-human oral (environmental) strains cocultivated in the laboratory. Clusters of host cell lysis by Ca. N. lyticus TM7x (bolded red) and Ca. M. amalyticus JR1 (bolded green) are represented more prominently with drawn lines.
Supplementary information
Supplementary Tables
Supplementary Table 1: list of putative genes involved in mycolic acid synthesis and export in CON44T, CON9T and seven additional Gordonia species. Supplementary Table 2: results of the REBASE restriction endonuclease analysis on six G. amarae and G. pseudoamarae strains. Putative restriction endonucleases identified are shown along with their closest match on the REBASE database, the type of restriction endonuclease system and predicted activity. Supplementary Table 3: results of the COG analysis of the 153 proteins (from the M. amalyticus genome) identified as a core Saccharibacteria proteome. Supplementary Table 4: full analyses of the T4SS genes found in M. amalyticus and an additional 25 ‘Candidatus Saccharibacteria’ genomes. This includes the composition and size of T4SS operons and BLASTp comparison of T4SS components in 25 ‘Candidatus Saccharibacteria’ genomes to homologues found in M. amalyticus. Supplementary Table 5: list of putative genes involved in metabolism and peptidoglycan synthesis in the M. amalyticus genome. Supplementary Table 6: list of the putative predatory arsenal of M. amalyticus, comprising identification of putative peptidases, lipases, glycanases and DNases/RNases found in the genome. Supplementary Table 7: results of the 16S rRNA gene amplicon analysis from previously published datasets. Wastewater treatment plants where M. amalyticus was positively identified are shown; for reference, the relative amount of ‘Candidatus S. aalborgensis’ (a Saccharibacteria species originally detected in wastewater treatment plants in Denmark) and five enriched bacterial genera (Tetrasphaera, Gordonia, Microthrix, Nocardioides and Terrimonas are shown. Supplementary Table 8: record of the NCBI GenBank accession numbers for genome sequences used throughout the study.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 1
Newick phylogenetic tree.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 1
Newick phylogenetic tree.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 5
Newick phylogenetic tree.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Newick phylogenetic tree.
Rights and permissions
About this article
Cite this article
Batinovic, S., Rose, J.J.A., Ratcliffe, J. et al. Cocultivation of an ultrasmall environmental parasitic bacterium with lytic ability against bacteria associated with wastewater foams. Nat Microbiol 6, 703–711 (2021). https://doi.org/10.1038/s41564-021-00892-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-021-00892-1
This article is cited by
-
Global abundance patterns, diversity, and ecology of Patescibacteria in wastewater treatment plants
Microbiome (2024)
-
Uncovering microbiomes of the rice phyllosphere using long-read metagenomic sequencing
Communications Biology (2024)
-
A novel and diverse group of Candidatus Patescibacteria from bathypelagic Lake Baikal revealed through long-read metagenomics
Environmental Microbiome (2023)
-
Vitamin interdependencies predicted by metagenomics-informed network analyses and validated in microbial community microcosms
Nature Communications (2023)
-
Bacteriophage therapy: an emerging paradigm in fish disease management
Aquaculture International (2023)