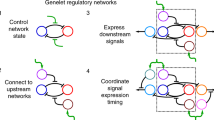
Abstract
Engineered far-from-equilibrium synthetic chemical networks that pulse or switch states in response to environmental signals could precisely regulate the kinetics of chemical synthesis or self-assembly. Currently, such networks must be extensively tuned to compensate for the different activities of and unintended reactions between a network’s various chemical components. Modular elements with standardized performance could be used to rapidly construct networks with designed functions. Here we develop standardized excitable chemical regulatory elements, termed genelets, and use them to construct complex in vitro transcriptional networks. We develop a protocol for identifying >15 interchangeable genelet elements with uniform performance and minimal crosstalk. These elements can be combined to engineer feedforward and feedback modules whose dynamics match those predicted by a simple kinetic model. Modules can then be rationally integrated and organized into networks that produce tunable temporal pulses and act as multistate switchable memories. Standardized genelet elements, and the workflow to identify more, should make engineering complex far-from-equilibrium chemical dynamics routine.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data associated with this manuscript are available at: https://doi.org/10.7281/T1/UBSZF1.
Code availability
The general genelet model code, including scripts for the main text simulations, is available at: https://github.com/sschaff6/general-genelet-model.git.
Change history
08 September 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41557-022-01058-0
References
Schultz, D., Wolynes, P. G., Jacob, E. B. & Onuchic, J. N. Deciding fate in adverse times: sporulation and competence in Bacillus subtilis. Proc. Natl Acad. Sci. USA 106, 21027–21034 (2009).
Oppenheim, A. B., Kobiler, O., Stavans, J., Court, D. L. & Adhya, S. Switches in bacteriophage lambda development. Annu. Rev. Genet. 39, 409–429 (2005).
Peter, I. S. & Davidson, E. H. Assessing regulatory information in developmental gene regulatory networks. Proc. Natl Acad. Sci. USA 114, 5862 (2017).
Alon, U. Network motifs: theory and experimental approaches. Nat. Rev. Genet. 8, 450–461 (2007).
van Esch, J. H., Klajn, R. & Otto, S. Chemical systems out of equilibrium. Chem. Soc. Rev. 46, 5474–5475 (2017).
van Roekel, H. W. H. et al. Programmable chemical reaction networks: emulating regulatory functions in living cells using a bottom-up approach. Chem. Soc. Rev. 44, 7465–7483 (2015).
Ferrell, J. E.Jr & Ha, S. H. Ultrasensitivity part III: cascades, bistable switches, and oscillators. Trends Biochem. Sci. 39, 612–618 (2014).
McAdams Harley, H. & Shapiro, L. Circuit simulation of genetic networks. Science 269, 650–656 (1995).
Ackermann, J., Wlotzka, B. & McCaskill, J. S. In vitro DNA-based predator–prey system with oscillatory kinetics. Bull. Math. Biol. 60, 329–354 (1998).
Montagne, K., Plasson, R., Sakai, Y., Fujii, T. & Rondelez, Y. Programming an in vitro DNA oscillator using a molecular networking strategy. Mol. Syst. Biol. 7, 466 (2011).
Semenov, S. N. et al. Rational design of functional and tunable oscillating enzymatic networks. Nat. Chem. 7, 160–165 (2015).
Kim, J. & Winfree, E. Synthetic in vitro transcriptional oscillators. Mol. Syst. Biol. 7, 465 (2011).
Montagne, K., Gines, G., Fujii, T. & Rondelez, Y. Boosting functionality of synthetic DNA circuits with tailored deactivation. Nat. Commun. 7, 13474 (2016).
Padirac, A., Fujii, T. & Rondelez, Y. Bottom-up construction of in vitro switchable memories. Proc. Natl Acad. Sci. USA 109, E3212–E3220 (2012).
Helwig, B., van Sluijs, B., Pogodaev, A. A., Postma, S. G. J. & Huck, W. T. S. Bottom-up construction of an adaptive enzymatic reaction network. Angew. Chem. Int. Ed. 57, 14065–14069 (2018).
Subsoontorn, P., Kim, J. & Winfree, E. Ensemble Bayesian analysis of bistability in a synthetic transcriptional switch. ACS Synth. Biol. 1, 299–316 (2012).
Postma, S. G. J., te Brinke, D., Vialshin, I. N., Wong, A. S. Y. & Huck, W. T. S. A trypsin-based bistable switch. Tetrahedron 73, 4896–4900 (2017).
Genot, A. J. et al. High-resolution mapping of bifurcations in nonlinear biochemical circuits. Nat. Chem. 8, 760 (2016).
Kim, J., White, K. S. & Winfree, E. Construction of an in vitro bistable circuit from synthetic transcriptional switches. Mol. Syst. Biol. 2, 68 (2006).
Kim, J., Khetarpal, I., Sen, S. & Murray, R. M. Synthetic circuit for exact adaptation and fold-change detection. Nucleic Acids Res. 42, 6078–6089 (2014).
Zadorin, A. S. et al. Synthesis and materialization of a reaction–diffusion French flag pattern. Nat. Chem. 9, 990 (2017).
Gines, G. et al. Microscopic agents programmed by DNA circuits. Nat. Nanotechnol. 12, 351 (2017).
Dupin, A. & Simmel, F. C. Signalling and differentiation in emulsion-based multi-compartmentalized in vitro gene circuits. Nat. Chem. 11, 32–39 (2019).
Green, L. N. et al. Autonomous dynamic control of DNA nanostructure self-assembly. Nat. Chem. 11, 510–520 (2019).
Franco, E. et al. Timing molecular motion and production with a synthetic transcriptional clock. Proc. Natl Acad. Sci. USA 108, E784–E793 (2011).
Meijer, L. H. H. et al. Hierarchical control of enzymatic actuators using DNA-based switchable memories. Nat. Commun. 8, 1117 (2017).
Schaffter, S. W. & Schulman, R. Building in vitro transcriptional regulatory networks by successively integrating multiple functional circuit modules. Nat. Chem. 11, 829–838 (2019).
Qian, L. & Winfree, E. Scaling up digital circuit computation with DNA strand displacement cascades. Science 332, 1196–1201 (2011).
Song, T. et al. Fast and compact DNA logic circuits based on single-stranded gates using strand-displacing polymerase. Nat. Nanotechnol. 14, 1075–1081 (2019).
Kishi, J. Y., Schaus, T. E., Gopalkrishnan, N., Xuan, F. & Yin, P. Programmable autonomous synthesis of single-stranded DNA. Nat. Chem. 10, 155–164 (2017).
Shah, S. et al. Using strand displacing polymerase to program chemical reaction networks. J. Am. Chem. Soc. 142, 9587–9593 (2020).
Chen, Z. et al. De novo design of protein logic gates. Science 368, 78 (2020).
Franco, E., Giordano, G., Forsberg, P.-O. & Murray, R. M. Negative autoregulation matches production and demand in synthetic transcriptional networks. ACS Synth. Biol. 3, 589–599 (2014).
Kim, J., Hopfield, J. & Winfree, E. in Advances in Neural Information Processing Systems 17 (eds Saul, L. K., Weiss, Y. & Bottou, L.) 681–688 (MIT Press, 2005).
Zadeh, J. N. et al. NUPACK: analysis and design of nucleic acid systems. J. Comput. Chem. 32, 170–173 (2011).
Dabby, N. Synthetic Molecular Machines for Active Self-assembly: Prototype Algorithms, Designs, and Experimental Study. PhD thesis, California Institute of Technology (2013).
Groves, B. et al. Computing in mammalian cells with nucleic acid strand exchange. Nat. Nanotechnol. 11, 287–294 (2016).
Isambert, H. The jerky and knotty dynamics of RNA. Methods 49, 189–196 (2009).
Zhang, D. Y. & Winfree, E. Control of DNA strand displacement kinetics using toehold exchange. J. Am. Chem. Soc. 131, 17303–17314 (2009).
Mangan, S. & Alon, U. Structure and function of the feed-forward loop network motif. Proc. Natl Acad. Sci. USA 100, 11980–11985 (2003).
Krupp, G. RNA synthesis: strategies for the use of bacteriophage RNA polymerases. Gene 72, 75–89 (1988).
Lapham, J. & Crothers, D. M. RNase H cleavage for processing of in vitro transcribed RNA for NMR studies and RNA ligation. RNA 2, 289–296 (1996).
Gardner, T. S., Cantor, C. R. & Collins, J. J. Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339–342 (2000).
Mahmoudabadi, G. & Phillips, R. A comprehensive and quantitative exploration of thousands of viral genomes. eLife 7, e31955 (2018).
O’Reilly, R. K., Turberfield, A. J. & Wilks, T. R. The evolution of DNA-templated synthesis as a tool for materials discovery. Acc. Chem. Res. 50, 2496–2509 (2017).
Schaffter, S. W. General Genelet Model (2020); https://github.com/sschaff6/general-genelet-model.git
Dubuc, E. et al. Cell-free microcompartmentalised transcription–translation for the prototyping of synthetic communication networks. Curr. Opin. Biotechnol. 58, 72–80 (2019).
Chatterjee, G., Dalchau, N., Muscat, R. A., Phillips, A. & Seelig, G. A spatially localized architecture for fast and modular DNA computing. Nat. Nanotechnol. 12, 920–927 (2017).
Laohakunakorn, N. et al. Bottom-up construction of complex biomolecular systems with cell-free synthetic biology. Front. Bioeng. Biotechnol. 8, 213 (2020).
Cunningham, P. & Ofengand, J. Use of inorganic pyrophosphatase to improve the yield of in vitro transcription reactions catalyzed by T7 RNA polymerase. BioTechniques 9, 713–714 (1990).
Acknowledgements
The authors thank E. Franco, E. Nakamura, M. Rubanov and P. Moerman for insightful conversations and comments on the manuscript. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship under grant number DGE-1232825 to S.W.S. This work was principally supported by the Department of Energy under award number DE-SC001 0426 to R.S. K.C. was supported by National Science Foundation award number EFRI-1830893 and Army Research Office award W911NF2010057. This work was also supported by the University of Chicago Materials Research Science and Engineering Center, which is funded by the National Science Foundation under award number DMR-2011854 to J.O. and A.M. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
S.W.S and R.S. designed the research. S.W.S conducted most of the experiments and simulations. K.C. performed the experiments presented in Supplementary Information, sections 11, 3.4 and 4.6. M.N. performed preliminary experiments for the study. J.O. and A.M. conducted the multistability simulations and analysis. S.W.S and R.S. wrote the paper with feedback from the other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Chemistry thanks James Carothers and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Information.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Schaffter, S.W., Chen, KL., O’Brien, J. et al. Standardized excitable elements for scalable engineering of far-from-equilibrium chemical networks. Nat. Chem. 14, 1224–1232 (2022). https://doi.org/10.1038/s41557-022-01001-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-022-01001-3
This article is cited by
-
DNA as a universal chemical substrate for computing and data storage
Nature Reviews Chemistry (2024)
-
Scaling up genelet circuits
Nature Chemistry (2022)