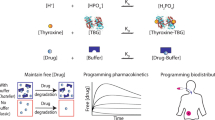
Abstract
Stimuli-responsive biomaterials that contain logic gates hold great potential for detecting and responding to pathological markers as part of clinical therapies. However, a major barrier is the lack of a generalized system that can be used to easily assemble different ligand-responsive units to form programmable nanodevices for advanced biocomputation. Here we develop a programmable polymer library by including responsive units in building blocks with similar structure and reactivity. Using these polymers, we have developed a series of smart nanocarriers with hierarchical structures containing logic gates linked to self-immolative motifs. Designed with disease biomarkers as inputs, our logic devices showed site-specific release of multiple therapeutics (including kinase inhibitors, drugs and short interfering RNA) in vitro and in vivo. We expect that this ‘plug and play’ platform will be expanded towards smart biomaterial engineering for therapeutic delivery, precision medicine, tissue engineering and stem cell therapy.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All the datasets generated and/or analysed during this study and supporting the findings described are available within the article and its Supplementary Information or from the corresponding author upon reasonable request.
References
Martinez-Outschoorn, U. E., Peiris-Pagés, M., Pestell, R. G., Sotgia, F. & Lisanti, M. P. Cancer metabolism: a therapeutic perspective. Nat. Rev. Clin. Oncol. 14, 11–31 (2016).
McKinney, E. F. & Smith, K. G. C. Metabolic exhaustion in infection, cancer and autoimmunity. Nat. Immunol. 19, 213–221 (2018).
Khan, A. T., Dobson, R. J. B., Sattlecker, M. & Kiddle, S. J. Alzheimer’s disease: are blood and brain markers related? A systematic review. Ann. Clin. Transl. Neurol. 3, 455–462 (2016).
Yu, T. C. et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 170, 548–563 (2017).
Nagarsheth, N., Wicha, M. S. & Zou, W. P. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat. Rev. Immunol. 17, 559–572 (2017).
Kalluri, R. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer 16, 582–598 (2016).
Wang, Y. F. & Kohane, D. S. External triggering and triggered targeting strategies for drug delivery. Nat. Rev. Mater. 2, 14 (2017).
Lu, Y., Aimetti, A. A., Langer, R. & Gu, Z. Bioresponsive materials. Nat. Rev. Mater. 1, 16075 (2016).
Ridker, P. M. et al. Effect of interleukin-1β inhibition with canakinumab on incident lung cancer in patients with atherosclerosis: exploratory results from a randomised, double-blind, placebo-controlled trial. Lancet 390, 1833–1842 (2017).
Gandhi, L. et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N. Engl. J. Med. 378, 2078–2092 (2018).
Mace, T. A. et al. IL-6 and PD-L1 antibody blockade combination therapy reduces tumour progression in murine models of pancreatic cancer. Gut 67, 320–332 (2018).
Li, J., Green, A. A., Yan, H. & Fan, C. H. Engineering nucleic acid structures for programmable molecular circuitry and intracellular biocomputation. Nat. Chem. 9, 1056–1067 (2017).
You, M. X., Zhu, G. Z., Chen, T., Donovan, M. J. & Tan, W. H. Programmable and multiparameter DNA-based logic platform for cancer recognition and targeted therapy. J. Am. Chem. Soc. 137, 667–674 (2015).
Peng, R. Z. et al. Engineering a 3D DNA-logic gate nanomachine for bispecific recognition and computing on target cell surfaces. J. Am. Chem. Soc. 140, 9793–9796 (2018).
Pugh, G. C., Burns, J. R. & Howorka, S. Comparing proteins and nucleic acids for next-generation biomolecular engineering. Nat. Rev. Chem. 2, 113–130 (2018).
Sedlmayer, F., Aubel, D. & Fussenegger, M. Synthetic gene circuits for the detection, elimination and prevention of disease. Nat. Biomed. Eng. 2, 399–415 (2018).
Chatterjee, G., Dalchau, N., Muscat, R. A., Phillips, A. & Seelig, G. A spatially localized architecture for fast and modular DNA computing. Nat. Nanotechnol. 12, 920–927 (2017).
Green, A. A. et al. Complex cellular logic computation using ribocomputing devices. Nature 548, 117–121 (2017).
Gao, X. J., Chong, L. S., Kim, M. S. & Elowitz, M. B. Programmable protein circuits in living cells. Science 361, 1252–1258 (2018).
Esabahy, M., Heo, G. S., Lim, S. M., Sun, G. R. & Wooley, K. L. Polymeric nanostructures for imaging and therapy. Chem. Rev. 115, 10967–11011 (2015).
Kamaly, N., Yameen, B., Wu, J. & Farokhzad, O. C. Degradable controlled-release polymers and polymeric nanoparticles: mechanisms of controlling drug release. Chem. Rev. 116, 2602–2663 (2016).
Zhang, Y. et al. Chain-shattering polymeric therapeutics with on-demand drug-release capability. Angew. Chem. Int. Ed. 52, 6435–6439 (2013).
Stuart, M. A. C. et al. Emerging applications of stimuli-responsive polymer materials. Nat. Mater. 9, 101–113 (2010).
Fu, X., Hosta-Rigau, L., Chandrawati, R. & Cui, J. Multi-stimuli-responsive polymer particles, films, and hydrogels for drug delivery. Chem 4, 2084–2107 (2018).
Ikeda, M. et al. Installing logic-gate responses to a variety of biological substances in supramolecular hydrogel–enzyme hybrids. Nat. Chem. 6, 511–518 (2014).
Badeau, B. A., Comerford, M. P., Arakawa, C. K., Shadish, J. A. & DeForest, C. A. Engineered modular biomaterial logic gates for environmentally triggered therapeutic delivery. Nat. Chem. 10, 251–258 (2018).
Liu, G. et al. Hyperbranched self-immolative polymers (hSIPs) for programmed payload delivery and ultrasensitive detection. J. Am. Chem. Soc. 137, 11645–11655 (2015).
Lei, E. K. & Kelley, S. O. Delivery and release of small-molecule probes in mitochondria using traceless linkers. J. Am. Chem. Soc. 139, 9455–9458 (2017).
Roth, M. E., Green, O., Gnaim, S. & Shabat, D. Dendritic, oligomeric, and polymeric self-immolative molecular amplification. Chem. Rev. 116, 1309–1352 (2016).
Staben, L. R. et al. Targeted drug delivery through the traceless release of tertiary and heteroaryl amines from antibody–drug conjugates. Nat. Chem. 8, 1112–1119 (2016).
Yakes, F. M. et al. Cabozantinib (XL184), a novel MET and VEGFR2 inhibitor, simultaneously suppresses metastasis, angiogenesis, and tumor growth. Mol. Cancer Ther. 10, 2298–2308 (2011).
Saatci, Ö. et al. Targeting PLK1 overcomes T-DM1 resistance via CDK1-dependent phosphorylation and inactivation of Bcl-2/xL in HER2-positive breast cancer. Oncogene 37, 2251–2269 (2018).
Wu, J., Ivanov, A. I., Fisher, P. B. & Fu, Z. Polo-like kinase 1 induces epithelial-to-mesenchymal transition and promotes epithelial cell motility by activating CRAF/ERK signaling. eLife 5, e10734 (2016).
Alouane, A., Labruère, R., Saux, T. L., Schmidt, F. & Jullien, L. Self-immolative spacers: kinetic aspects, structure–property relationships, and applications. Angew. Chem. Int. Ed. 54, 7492–7509 (2015).
Zhang, D. et al. Linker immolation determines cell killing activity of disulfide-linked pyrrolobenzodiazepine antibody–drug conjugates. ACS Med. Chem. Lett. 7, 988–993 (2016).
Yueqin, Z. et al. Esterase-sensitive prodrugs with tunable release rates and direct generation of hydrogen sulfide. Angew. Chem. Int. Ed. 55, 4514–4518 (2016).
Wu, S. & Fu, L. Tyrosine kinase inhibitors enhanced the efficacy of conventional chemotherapeutic agent in multidrug resistant cancer cells. Mol. Cancer 17, 25 (2018).
Fu, Z. & Wen, D. The emerging role of polo-like kinase 1 in epithelial-mesenchymal transition and tumor metastasis. Cancers 9, 131 (2017).
Narvaez, A. J. et al. Modulating protein-protein interactions of the mitotic polo-like kinases to target mutant KRAS. Cell Chem. Biol. 24, 1017–1028 (2017).
Yun, J. et al. Vitamin C selectively kills KRAS and BRAF mutant colorectal cancer cells by targeting GAPDH. Science 350, 1391–1396 (2015).
Yang, B. W., Chen, Y. & Shi, J. L. Reactive oxygen species (ROS)-based nanomedicine. Chem. Rev. 119, 4881–4985 (2019).
Sullivan, L. B., Gui, D. Y. & Heiden, M. G. V. Altered metabolite levels in cancer: implications for tumour biology and cancer therapy. Nat. Rev. Cancer 16, 680–693 (2016).
Vander-Heiden, M. G. & DeBerardinis, R. J. Understanding the intersections between metabolism and cancer biology. Cell 168, 657–669 (2017).
Ghandi, M. et al. Next-generation characterization of the cancer cell line encyclopedia. Nature 569, 503–508 (2019).
Li, H. et al. The landscape of cancer cell line metabolism. Nat. Med. 25, 850–860 (2019).
Webb, B. A., Chimenti, M., Jacobson, M. P. & Barber, D. L. Dysregulated pH: a perfect storm for cancer progression. Nat. Rev. Cancer 11, 671–677 (2011).
Sabharwal, S. S. & Schumacker, P. T. Mitochondrial ROS in cancer: initiators, amplifiers or an Achilles’ heel? Nat. Rev. Cancer 14, 709–721 (2014).
Tang, Z. F. et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45, W98–W102 (2017).
Holohan, C., Van Schaeybroeck, S., Longley, D. B. & Johnston, P. G. Cancer drug resistance: an evolving paradigm. Nat. Rev. Cancer 13, 714–726 (2013).
Aron, A. T. et al. In vivo bioluminescence imaging of labile iron accumulation in a murine model of Acinetobacter baumannii infection. Proc. Natl Acad. Sci. USA 114, 12669–12674 (2017).
Bruemmer, K. J., Green, O., Su, T. A., Shabat, D. & Chang, C. J. Chemiluminescent probes for activity-based sensing of formaldehyde released from folate degradation in living mice. Angew. Chem. Int. Ed. 57, 7508–7512 (2018).
Gorrini, C., Harris, I. S. & Mak, T. W. Modulation of oxidative stress as an anticancer strategy. Nat. Rev. Drug Discov. 12, 931–947 (2013).
Steeg, P. S. Targeting metastasis. Nat. Rev. Cancer 16, 201–218 (2016).
Gotwals, P. et al. Prospects for combining targeted and conventional cancer therapy with immunotherapy. Nat. Rev. Cancer 17, 286–301 (2017).
Spring, B. Q. et al. A photoactivable multi-inhibitor nanoliposome for tumour control and simultaneous inhibition of treatment escape pathways. Nat. Nanotechnol. 11, 378–387 (2016).
Fan, W., Yung, B., Huang, P. & Chen, X. Nanotechnology for multimodal synergistic cancer therapy. Chem. Rev. 117, 13566–13638 (2017).
Chen, Z. H. et al. Targeting genomic rearrangements in tumor cells through Cas9-mediated insertion of a suicide gene. Nat. Biotechnol. 35, 543 (2017).
Acknowledgements
This research was supported by the National Natural Science Foundation of China (21605118, 21974105, 11522219, 11972280, 11532009, 21427807, 21834004, 21827811 and 61527806), the China Postdoctoral Science Foundation (2015M580829 and 2017T100735), the Postdoctoral Science Foundation of Shaanxi Province (2016BSHTBSHTDZZ01), the Natural Science Foundation of Shaanxi Province (2018JQ2001), the Open Funds of SKLACLS from Nanjing University (1508) and the Fundamental Research Funds for the Central Universities (xjj2015081). Z.P. also acknowledges financial support from the program of China Scholarships Council. J.Z. thanks the support from Excellent Research Program of Nanjing University (ZYJH004). Correspondence and requests for materials should be addressed to W.T., F.X. or J.-J.Z.
Author information
Authors and Affiliations
Contributions
P.Z., J.-J.Z., F.X. and W.T. conceived the project, analysed the data and discussed the writing of the paper. P.Z., D.G., K.A., Q.S., Y.Z., X.C., X.D. and J.L. designed the monomers and performed the organic synthesis experiments. P.Z., K.A., C.W., X.P., T.L., C.C., Y.L. and X.H. performed the in vitro and in vivo experiments and analysed the data. T.Y. analysed the gene expression levels from the CCLE and TCGA databases. P.Z., W.T. and F.X. wrote the manuscript. P.Z., D.G. and K.A. contributed equally to this work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Methods, additional discussion, Supplementary Figs. 1–20, Tables 1–4, profiles of gene expression, and references.
Rights and permissions
About this article
Cite this article
Zhang, P., Gao, D., An, K. et al. A programmable polymer library that enables the construction of stimuli-responsive nanocarriers containing logic gates. Nat. Chem. 12, 381–390 (2020). https://doi.org/10.1038/s41557-020-0426-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-020-0426-3
This article is cited by
-
Dual-controlled guest release from coordination cages
Communications Chemistry (2024)
-
Bionic natural small molecule co-assemblies towards targeted and synergistic Chemo/PDT/CDT
Biomaterials Research (2023)
-
Emulsion-oriented assembly for Janus double-spherical mesoporous nanoparticles as biological logic gates
Nature Chemistry (2023)
-
Bioinspired oral delivery devices
Nature Reviews Bioengineering (2023)
-
Ratiometric afterglow luminescent nanoplatform enables reliable quantification and molecular imaging
Nature Communications (2022)