Abstract
We aim to elucidate the prognostic value of PIK3CA mutations and copy number (CN) gain (PIK3CA-mut/gain) in hormone receptor-positive and HER2-negative (HR + /HER2−) breast cancer (BC). We analyzed primary HR + /HER2− BC from three publicly available datasets comprising over 2000 samples and assessed the associations with tumoral and clinical characteristics and outcome. Clinical benefit (CB) in alpelisib-treated patients from two studies including 46 patients was analyzed. About 8–10% of HR + /HER2− primary BC had PIK3CA-mut/gain. In two of the datasets analyzed, among patients with PIK3CA mutant tumors, those with mut/gain had significantly worse outcome compared to those with CN neutral (PIK3CA-mut/neut) and PIK3CA-mut/gain remained an independent prognostic factor. CB of alpelisib-treated patients with PIK3CA-mut/gain and PIK3CA-mut/neut tumors was comparable. PIK3CA CN might help clarifying the prognostic and predictive role of PIK3CA mutations. Further studies are warranted.
Similar content being viewed by others
Introduction
The phosphoinositide 3-kinases (PI3K) pathway plays a critical role in breast cancer (BC) and is frequently altered in hormone receptor positive and HER2 negative (HR + /HER2−) disease1. Somatic mutations of the PIK3CA gene, encoding for the class IA PI3K p110α subunit, are the most common activating mutations, occurring in 30–50% of ER + /HER2− early BC2,3 and in 28% of metastatic disease4. Many studies evaluated the prognostic relevance of PIK3CA mutations in primary BC with conflicting results2,5,6. The approval of alpelisib, a selective PI3K-alpha inhibitor, for the treatment of patients with PIK3CA mutant HR + /HER2− advanced BC progressing on prior endocrine therapies7 brought to a renewed interest in PIK3CA as predictive marker in HR + /HER2− BC.
Gain in PIK3CA copy number (CN) has been described in BC8,9,10,11,12,13,14,15,16,17,18. It was shown that tumors with high PIK3CA CN have more aggressive prognostic features, including large tumor size, high tumor grade, and negative HR status and are more likely to occur in patients with HR and HER2 negative disease9. In about half of the tumors, gain in PIK3CA CN co-occurs with PIK3CA mutations8,10.
Despite the overwhelming number of studies assessing the prognostic and predictive role of PIK3CA mutations, a comprehensive study combining PIK3CA mutations and CN in HR + /HER2− BC is lacking.
In this study, we aimed to perform a combined analysis of PIK3CA mutations and CN gain in three large and well characterized BC cohorts, namely METABRIC19,20, MSK-breast cancer 2018 (MSK-2018)21 and TCGA-BRCA (TCGA)22. In addition, we aimed to gain insights on the role of PIK3CA gain as a potential predictive marker of response to alpelisib in publicly available datasets of cancer cell lines23, patients derived xenograft (PDX)24 and patients with metastatic BC25,26,27.
Results
PIK3CA genomic alterations are associated with tumoral and clinical characteristics in HR + /HER2- BC
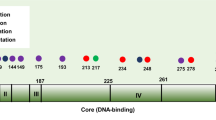
Gain in PIK3CA CN was observed in 194/1377 (14.1%) of HR + /HER2− primary BC within METABRIC. Tumors with gain in PIK3CA showed a significant increase in PIK3CA mRNA expression compared to PIK3CA neutral (p = 1.6e−05) (Fig. 1a), even when only PIK3CA mutant tumors were considered (p = 8.5e−05) (Fig. 1b). Gain in PIK3CA occurred more frequently in PIK3CA mutant compared to wild-type (wt) tumors (18.2% versus 10.6% p = 8.3e−05) (Fig. 1c). When mutated, tumors with PIK3CA gain had a similar proportion of mutations in exons 10 and 21 (p = 0.89), hot-spot mutations (p = 0.34) and double mutations (p = 0.6) compared to PIK3CA neutral (Fig. 1d–f, respectively). These analyses were also performed in luminal A and luminal B BC separately, with similar results (Supplementary Fig. 1).
Distribution of PIK3CA mRNA according to PIK3CA categories (a, b); PIK3CA gain according to mutational status (c); mutation exons (d), hotspots (e) and double mutations (f) according to PIK3CA gain status. Analyses were performed in HR + /HER2− BC within METABRIC using Mann–Whitney–Wilcoxon in a, b and Two-proportion z-test in c–f. Levels of PIK3CA log2ratio in primary and metastatic HR + /HER2− BC within MSK-2018 (g). Mann–Whitney–Wilcoxon test was performed. For box plots, lower and upper bars correspond to the minimum and maximum non-outlier values of the data distribution. Outliers are defined as values outside of the range (Q1 − 1.5×(Q3 − Q1), Q3 + 1.5×(Q3 − Q1)), where Q1 and Q3 are the first and third quartile, respectively.
In MSK-2018, significantly higher levels of PIK3CA CN were observed in metastatic HR + /HER2− compared to primary BC samples, both in tumors unselected for PIK3CA mutations (p = 5.3e−24), and in PIK3CA mutant and wt tumors (Fig. 1g and Supplementary Fig. 2). However, the proportion of samples with both PIK3CA mutations and CN gain were not significantly different between the de-novo and not de-novo metastatic groups (p = 0.17, Supplementary Fig. 2).
Table 1 and Supplementary Tables 1 and 2 show the clinico-pathological characteristics of HR + /HER2− patients within METABRIC, MSK-2018 and TCGA, respectively, according to CN gain and mutational status of PIK3CA. Four categories were evaluated: PIK3CA wt and CN neutral (-wt/neut), PIK3CA wt with CN gain (-wt/gain), PIK3CA mutant and CN neutral (-mut/neut), and PIK3CA mutant with CN gain (-mut/gain). PIK3CA-mut/gain was observed in 8.3%, 7% and 10% of patients within METABRIC, MSK-2018 and TCGA, respectively. In all datasets PIK3CA categories were significantly associated with the histological subtypes. In METABRIC and MSK-2018 a significant association with grade was found. In METABRIC and TCGA significant associations with size and luminal subtypes were also observed. Nodal status and the Integrative Clusters (IC) based on copy number alterations (CNA)19 were significantly associated with PIK3CA categories in METABRIC. Tumors with PIK3CA-gain (both PIK3CA-wt/gain and PIK3CA-mut/gain) were more frequently of higher grade, larger than 2 cm and luminal B. PIK3CA-mut/neut tumors were more frequently lobular or mixed and luminal A. PIK3CA-mut/gain was observed with higher frequency within IC 3 and 8, while PIK3CA-wt/gain was more frequent in IC 1 and 9.
In all datasets we aimed to establish if there were genomic mutations enriched in PIK3CA-mut/gain tumors compared to PIK3CA-mut/neut and found that TP53 mutations were indeed significantly enriched (q value < 0.05) in primary PIK3CA-mut/gain BC samples from METABRIC and MSK-2018 while only a borderline significant association was found in TCGA (q value = 0.06). In METABRIC we also found an enrichment of SF3B1 mutations in PIK3CA-mut/gain and of GATA3 mutations in PIK3CA-mut/neut BC. A complete list of the mutations analyzed in all datasets is reported as supplementary table 3.
PIK3CA-mut/gain is significantly and independently associated with outcome in HR + /HER2− BC
In METABRIC, when comparing the outcome of patients with PIK3CA-mut/gain versus those with PIK3CA-mut/neut tumors, we found a significantly worse recurrence-free (RFS) (p = 0.0055) and disease-specific survival (DSS) (p = 0.0026) for the PIK3CA-mut/gain group, in both unselected patients and in those with luminal A (RFS p = 0.042, DSS p = 0.07) but not luminal B BC (RFS p = 0.29, DSS p = 0.29) (Fig. 2a–c and Supplementary Fig. 3a–c). This was probably related to the different prognostic role of PIK3CA mutations observed in patients without PIK3CA gain in luminal A versus B subtypes. Indeed, in patients with luminal A BC there was no significant difference in terms of RFS (p = 0.63) or DSS (p = 0.68) between patients with PIK3CA-mut/neut or PIK3CA-wt/neut tumors, while in patients with luminal B BC a worse RFS and DSS was found for those with PIK3CA-mut/neut compared to PIK3CA-wt/neut, even though results were statistically significant only for DSS (p = 0.02; RFS p = 0.11) (Fig. 2b, c and Supplementary Fig. 3b, c).
Kaplan–Meier curves of RFS according to the four categories of PIK3CA in all patients with HR + /HER2− BC (a) or in those with luminal A (b) or B (c) BC within METABRIC and Kaplan–Meier curves of DFS in all patients with primary HR + /HER2− BC within MSK-2018 (d). For each category, the number of patients at risk is indicated.
In MSK-2018, consistently with data from METABRIC, we observed a significantly worse disease-free survival (DFS) (p = 0.00062) for patients with PIK3CA-mut/gain compared to PIK3CA-mut/neut with overall survival (OS) data only showing a trend for significance (p = 0.084) (Fig. 2d and supplementary Fig. 3d).
In TCGA we were unable to confirm the significant association with outcome in patients with HR + /HER2− PIK3CA-mut/gain BC (p = 0.48), despite the significant associations with poor prognostic factors.
We also analyzed the prognostic value of PIK3CA mut/gain in patients with primary HR + /HER2− BC receiving adjuvant endocrine therapy. A significantly worse survival for patients with PIK3CA-mut/gain tumors compared to those with PIK3CA-mut/neut, in both METABRIC (RFS p = 0.0034) and MSK-2018 (DFS p = 0.0036) was observed, confirming the poor prognostic role of PIK3CA-mut/gain in patients receiving endocrine therapy (supplementary Fig. 4a, b). In addition, we found a higher proportion of PIK3CA-mut/gain tumors in patients receiving endocrine therapy who relapsed compared to those who did not relapse (supplementary Fig. 4c).
In METABRIC and MSK-2018 we performed multivariate analyses, taking into account age, grade, size, nodal status and histological subtypes and the four categories of PIK3CA. PIK3CA-mut/gain maintained an independent prognostic role for both RFS (p = 0.015) and DSS (p = 0.012) in METABRIC, and for DFS (p = 0.023) in MSK-2018 (Fig. 3 and supplementary Fig. 5).
PIK3CA-mut/gain does not seem to provide additional informations for alpelisib response in patients with HR + /HER2− BC
We first analyzed cancer cell lines with available IC50 data on alpelisib and PIK3CA mutational and CNA data23. As expected, both BC and pan-cancer PIK3CA-mut/neut cell lines showed significantly lower IC50 values compared to PIK3CA-wt/neut (p = 0.0059 and p = 1.1e−02, respectively) while pan-cancer, but not BC cells with PIK3CA-mut/gain showed significantly lower IC50 values compared to PIK3CA-mut/neut (p = 0.016 and p = 0.95, respectively) (Fig. 4a and supplementary Fig. 6a).
Box-plots of alpelisib IC50 data in BC cell lines (a) and change in tumor volume in BC PDX treated with alpelisib (b) according to the PIK3CA categories. Bar-plots of month on treatment for alpelisib-treated patients within ALP-201926,27 according to the PIK3CA categories (c). Bar-plots of weeks on study for alpelisib-treated patients within ALP-202025 according to the PIK3CA categories (d); Mann–Whitney–Wilcoxon tests were performed in a and b. For box plots, lower and upper bars correspond to the minimum and maximum non-outlier values of the data distribution. Outliers are defined as values outside of the range (Q1 − 1.5×(Q3 − Q1), Q3 + 1.5×(Q3 − Q1)), where Q1 and Q3 are the first and third quartile, respectively.
We next analyzed the responses to alpelisib in a large and well characterized dataset of PDX24. Responses to alpelisib were not significantly different when analyzing only BC PDX (Fig. 4b); however, when considering pan-cancer PDX we observed significantly better responses in tumors with PIK3CA-mut/gain compared to PIK3CA-mut/neut (p = 0.023) (supplementary Fig. 6b).
We finally analyzed patients with ER + /HER2− BC treated with alpelisib and endocrine therapy for metastatic disease included in two different datasets, that were termed ALP-201926,27 and ALP-202025. In ALP-2019, 12 patients received alpelisib and letrozole26,27. Among these, three had PIK3CA-mut/gain tumors and two of these patients (67%) derived CB; CB rate for patients with PIK3CA-mut/neut was 50% (Fig. 4c). In ALP-2020, 34 patients received alpelisib in combination with letrozole or exemestane25. Among the 7 patients with PIK3CA-mut/gain tumors, 4 (57%) derived CB. CB was observed in 62% of patients with PIK3CA-mut/neut tumors (Fig. 4d).
Discussion
In this study we primarily aimed to perform a comprehensive analysis on the prognostic role of PIK3CA CN gain with co-occurring PIK3CA mutations in well characterized and publicly available datasets of patients with HR + /HER2− BC.
Previous studies have documented the gain in PIK3CA CN in patients with BC, but reports on its frequency have been conflicting, ranging from 1.4%13 to as high as 72%14, with two of the most recent studies reporting frequencies of 9%9 and 17.4%15 in HR + and luminal/HER2−, respectively. PIK3CA CN has been explored by polymerase chain reaction (PCR)10,11,12,13,14,16,17, single nucleotide polymorphism (SNP) array8,9 and next generation sequencing (NGS)15 and different cut-offs and definitions (PIK3CA gain versus amplification) have been used, potentially explaining the wide and discrepant ranges in PIK3CA gain frequency. We detected gain in PIK3CA CN in 14.1% of HR+/HER2− primary BC in METABRIC, which is in line with two of the most recent reports9,15. In the present study we considered together tumors with PIK3CA gain and amplification (DNAcopy status 1 and 2, respectively in METABRIC). When analyzing PIK3CA mRNA expression in tumors with PIK3CA gain and amplification, we found a significantly higher PIK3CA mRNA expression for amplified tumors compared to those with gain (p = 0.00017), as expected, yet significantly higher mRNA levels were observed in PIK3CA gain versus neutral tumors (p = 0.02) (supplementary Fig. 7a). Additionally, among patients with PIK3CA mutant tumors, a significantly worse RFS was observed for patients with PIK3CA amplification or PIK3CA gain (supplementary Fig. 7b), indicating that, in patient with PIK3CA mutant tumors, PIK3CA gain might have clinical relevance in addition to PIK3CA amplification.
We observed that PIK3CA CN gain occurred preferentially in PIK3CA mutant tumors, in accordance with previous reports8 and supporting the hypothesis of a potential additive effect of mutations and gain to oncogenesis8. Mutations in the helical and kinase domain of PIK3CA have been previously associated with different outcome in patients with BC28 and double mutations were shown to induce increased PI3K activity and signaling and increased tumor proliferation29. In our study, differently from Kadota et al.8 we did not find a significant association between gain in PIK3CA and any PIK3CA mutation exons nor we found any association with PIK3CA double mutations or hotspots mutations. Interestingly, we observed a significant increase in PIK3CA CN in metastatic compared to primary tumors in MSK-2018, which might suggest a potential role for PIK3CA CN in the metastatic process. However, a correction for cellularity or other confounding factors was not performed, therefore caution must be taken in interpreting this data.
It has been previously demonstrated a significant association between PIK3CA CN and high grade, stage and HR- status in an unselected population with BC9. Here we demonstrated the significant associations with grade, size and nodal status also in patients with HR+ /HER2− BC. In addition, we found a significant association with luminal subtypes and, accordingly, with the histological subtypes. The significant association with TP53 mutations is also coherent with these findings.
When we analyzed survival according to the PIK3CA categories derived from the combination of CN and mutations, a significantly worse outcome was observed in patients with PIK3CA-mut/gain compared to -mut/neut tumors in METABRIC and MSK-2018. Of note, the prognostic role of PIK3CA-mut/gain was independent of grade, size, histological subtype and nodal status in both datasets. We also found that PIK3CA-mut/gain was prognostic in patients receiving endocrine therapy and that patients relapsing during endocrine therapy had more frequently PIK3CA-mut/gain tumors. Whether PIK3CA-mut/gain status might be associated with endocrine resistance should be better evaluated in future studies. Prior to our study, the prognostic relevance of PIK3CA CN has been demonstrated in pan-cancer studies30,31, but in patients with primary HR + BC one of the largest studies failed to establish an association between PIK3CA CN and outcome9. In previous studies the combined evaluation of PIK3CA gain and mutations was not performed. Our results suggest that assessing PIK3CA gain together with PIK3CA mutations might give a better estimation of the prognostic value of PIK3CA in patients with HR + /HER2− BC.
An interesting observation in our study was the different effect on outcome of PIK3CA mutations and gain in patients with luminal A and luminal B BC. Compared to patients with PIK3CA-mut/neut, those with PIK3CA-mut/gain luminal A BC experienced worse RFS. This was not observed in luminal B BC, where patients with PIK3CA-mut/neut tumors showed a worse outcome compared to PIK3CA-wt/neut. We have not thoroughly investigated the potential explanations of these observations. We analyzed whether PIK3CA cancer cell fraction, the DNAcopy status, the presence of double mutations or a different proportion of mutation exons were associated with luminal subtypes. However, no significant differences were found (supplementary Fig. 8). Pereira et al. previously demonstrated that PIK3CA mutations have distinct prognostic associations in ER + tumors stratified into IC20, and some of these have different proportion of luminal A and luminal B subtypes19. Analysing 861 BC samples, Wilson TR et al. showed that patients with PIK3CA mutant luminal A BC tended to show a favorable but not statistically significant DFS32. This effect was not observed in patients with luminal B BC32. It was recently shown that patients with PIK3CA mutant luminal A BC were more likely to derive CB from PI3K inhibitors (alpelisib and buparlisib) compared to those with luminal B26. Based on ours and previous data it could be hypothesized that PIK3CA exerts its effects in a context-dependent manner, but this needs to be tested in future studies. Data regarding the prognostic role of PIK3CA mutations in HR +/HER2− BC have been controversial1,2,5. Whether different proportion of the luminal subtypes and PIK3CA gain might explain the different associations between PIK3CA mutations and outcome observed in previous studies remains a hypothesis. Nevertheless, our data support the evaluation of molecular subtypes and PIK3CA CN when assessing the prognostic role of PIK3CA.
In our study we also aimed to investigate whether a classification based on both PIK3CA gain and mutations could help clarifying the predictive role of PIK3CA as a marker of alpelisib response. The evidence that double PIK3CA mutations results in increased sensitivity to PI3Kα inhibitors compared with single-hotspot mutations29 could suggest that multiple hits on PIK3CA might have a synergistic effect. In our study better responses to alpelisib were observed in pan-cancer but not BC cell lines and PDX with PIK3CA-mut/gain compared to -mut/neut, probably due to the limited sample size. Additionally, patients receiving alpelisib with PIK3CA-mut/gain tumors do not seem to show different CB compared to those with PIK3CA-mut/neut tumors, which might suggest that response to alpelisib mainly depend upon the PIK3CA mutational rather than PIK3CA CN status. However, given the limited number of alpelisib-treated patients analyzed in our study, whether PIK3CA-mut/gain might predict different sensitivity to PI3K inhibitors needs to be established in larger studies. Also, further studies are needed to clarify if patients with HR +/HER2− tumors with PIK3CA-wt/gain might benefit from PI3K inhibitors. Indeed, in BC cells a lower although not statistically significant IC50 for alpelisib was observed, but among patients treated with alpelisib none had PIK3CA-wt/gain tumors. Overall, our results encourage the further combined evaluation of PIK3CA gain and mutations as a marker of PI3K inhibitors response.
We are aware of the limitations of our study. First, the analyses were retrospective and were performed in very heterogenous populations. Second, comparing CN alteration calls from different datasets is challenging because of the different methodologies and computational approaches used to generate these data. In particular, in METABRIC we used the DNAcopy data, in TCGA we used GISTIC 2.0 data, while in MSK-2018, ALP-2019 and ALP-2020 we utilized log2-ratios data and identified an arbitrary, albeit data-driven, cut-off of PIK3CA based on the frequencies observed in METABRIC. We are aware that a univocal and clinically relevant cut-off remains to be set in future studies. For the same reason we did not analyze the enrichment/depletion of CN alterations differentiating PIK3CA-mut/gain and mut/neut BC, as done for mutations. Third, data on the predictive role of PIK3CA-mut/gain to alpelisib from BC and pan-cancer cells and PDX are not univocal and data from patients treated with alpelisib derive from very limited and non-randomized cohorts. Therefore, results are far to be considered conclusive. Further evidence on the predictive role of PIK3CA-mut/gain is needed from randomized clinical trials. Fourth, experimental approaches are needed to elucidate the mechanisms by which PIK3CA gain and PIK3CA mutations cooperate in inducing worse outcome and a differential effect in luminal subtypes. As other genes may be co-amplified with PIK3CA33, it would be interesting to investigate if any of these genes might have a role in the development of an aggressive phenotype in addition to PIK3CA.
On the other hand, our data were generated from three large, well characterized cohorts of BC and the poor outcome of patients with PIK3CA-mut/gain BC was replicated independently in two of the datasets. We made very interesting and thought-provoking observations: first, patients with HR + /HER2− BC with PIK3CA-gain/mut have worse outcome, independently of the most relevant clinico-pathological characteristics; second, PIK3CA mutations and CN gain might hold different prognostic effects in luminal A and luminal B BC; third, although very preliminary, our data from pan-cancer cell lines and PDX suggest that response to alpelisib might be influenced by PIK3CA CN gain.
In conclusion our data suggest that taking into account PIK3CA CN in addition to mutations might bring to a better evaluation of the PI3K pathway and help elucidating some controversial issues regarding the prognostic and predictive role of PIK3CA. Given the central role of PI3K pathway in tumor biology, outcome and prediction to therapy in patients with HR + /HER2− BC, further studies evaluating the combined effect of PIK3CA gain and mutations are warranted.
Methods
Datasets and data collection
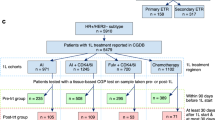
For METABRIC19,20, genomic, transcriptomic, clinical and outcome data of 2509 primary tumor samples from patients with BC were downloaded from CBioPortal34,35 (http://cbioportal.org) and patients with HR + /HER2− BC (n = 1413) were selected. PIK3CA protein-affecting mutations and CNA based on DNAcopy36 were considered. Data on mutated PIK3CA exons were downloaded from http://github.com/cclab-brca.
For MSK-201821, genomic, clinical and outcome data of 918 primary and 1000 metastatic tumor samples from 1715 patients with BC were accessed via CBioPortal34,35. PIK3CA protein-affecting mutations and CNA data based on log2-ratio profiles of HR + /HER2− BC (n = 1365) were considered for downstream analyses. Additional clinical data including treatment and de-novo metastatic status were downloaded from the supplementary materials of the original manuscript21.
For TCGA22, genomic, clinical and outcome data of 1084 primary tumor samples were downloaded from cBioPortal34,35. Additional clinical data on patients’ receptor status were downloaded from https://gdc.cancer.gov/access-data/gdc-data-portal, by means of TCGAbiolinks R/Bioconductor package (https://bioconductor.org/packages/release/bioc/html/TCGAbiolinks.html). Patients with HR + /HER2− BC (n = 440) were selected, and PIK3CA protein-affecting mutations and CNA based on GISTIC 2.037 were considered.
For ALP-201926,27, genomic, clinical and outcome data of 70 primary and metastatic samples from 68 ER + /HER2− patients were downloaded from CBioPortal34,35. Based on treatment data (downloaded from the supplementary materials from the original manuscript26,27), 12 alpelisib-treated patients were selected for the downstream analysis, along with PIK3CA mutational and CNA status based on log2-ratio genomic profiles.
For ALP-202025, genomic, clinical and outcome data for 51 primary and metastatic tumor samples from 51 HR + /HER2− patients treated with alpelisib were downloaded from CBioPortal34,35 and PIK3CA mutational and CNA status based on log2-ratio were considered for downstream analyses.
For Genomics of Drug Sensitivity in Cancer (GDSC) cell lines23, drug data for the PI3K/mTOR inhibitor alpelisib, response data and genetic features of 50 BC and 765 pan-cancer cell lines were downloaded from https://www.cancerrxgene.org/. PIK3CA mutational status, CNA data based on GISTIC and drugs IC50 were considered for downstream analyses.
For Novartis Institutes for BioMedical Research (NIBR) PDXE24, genomic information, treatment and response data of 277 PDX models across 6 tumor types (BC (n = 43), cutaneous melanoma (n = 33), colorectal carcinoma (n = 59), gastric cancer (n = 64), non-small cell lung carcinoma (n = 36) and pancreatic ductal adenocarcinoma (n = 42)) were retrieved from the original publication24. PDX models treated with alpelisib were selected, and PIK3CA mutational status, CNA status, treatment and response data were considered for downstream analyses.
Definition of CNA gain events
For METABRIC, PIK3CA CN gains and losses were defined based on DNAcopy calls36. Cases with PIK3CA CN loss were excluded, leaving 1377 patients for downstream analyses. In MSK-2018, PIK3CA CN gains and losses were defined applying the percentile of CN gain and loss events observed in HR + /HER2− patients from METABRIC (0.15 and 0.05 respectively) to the PIK3CA log2-ratio values of primary HR + /HER2− samples. As a result, PIK3CA log2-ratio greater than 0.1 were considered as CN gain and log2-ratio lower than -0.27 were considered as CN loss events. Cases with PIK3CA CN loss were excluded from downstream analysis. The same thresholds were applied to the ALP-2019 and ALP-2020 datasets.
For TCGA PIK3CA CN gains and losses were defined based on GISTIC 2.0 calls37. After the exclusion of cases with PIK3CA CN loss, 413 patients were considered for downstream analyses.
For GDSC cell lines, CN gains and losses were defined based on GISTIC calls. For PDXE, CNA calls from ExomeCNV were used to define PIK3CA gain and loss events.
Genomic analyses
The lists of the enriched mutations in METABRIC, MSK-2018 and TCGA were generated through cBioPortal34,35 by comparing HR + /HER2− BC samples categorized as PIK3CA-mut/gain and PIK3CA-mut/neut. In MSK-2018, primary and metastatic BC samples were analyzed separately and metastatic samples with unconfirmed HR + /HER2− biopsy status were excluded. As genomic mutations from these datasets were generated using different gene panels, we focused on a list of relevant BC genes from IntOGen (October 2021, n = 99)38. To make enrichment analyses comparable across the three datasets, statistically significant genes were identified using a restricted hypothesis testing (RHT) analysis. For each dataset, the p-values estimated from the cBioPortal analysis were adjusted by a Benjamini-Hochberg (BH) correction considering the list of relevant BC genes included in the dataset.
Statistical and survival analyses
Statistical/association analyses, data processing and plots were performed using the R environment (R Core Team, http://cran.r-project.org/). Mann–Whitney–Wilcoxon test was used to check for significant shifts between two distributions. Two-proportion z-test was used to compare proportions in two groups. Fisher’s exact test was used for comparison between two categorical variables. Tests were performed two-sided. Kaplan–Meier curves and log-rank test were used for all survival analyses. Cox proportional hazards model was used for multivariate analyses, including as covariates PIK3CA status, age, tumor grade, tumor size, histological subtypes and lymph-node status.
For METABRIC and MSK-2018 clinical endpoints were retrieved from cBioPortal: DSS and RFS for METABRIC, OS and DFS for MSK-2018. For ALP-2019, patients were considered to achieve CB when showed a stable disease for more than 6 months, in accordance with26; while for ALP-2020, patients were considered to achieve CB when showed a stable disease for more than 16 weeks, as in25. For GDSC cell lines, drug-specific IC50 values were downloaded from https://www.cancerrxgene.org/. For NIBR PDXE, best average response to alpelisib (change in tumor volume %) was considered24.
Within MSK-2018, patients with synchronous primary tumors and de novo metastatic were excluded from analyses involving primary samples; to avoid duplicates, when more than one sample was present, the treatment naïve one was chosen. When duplicated samples could not be solved, cases were excluded. For survival analysis in patients treated with endocrine therapy, patients with primary HR + /HER2− BC treated with hormonal adjuvant systemic therapy were selected.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Code availability
Codes used to generate the data are available upon reasonable request.
References
Criscitiello, C., Marra, A. & Curigliano, G. PIK3CA mutation assessment in HR+/HER2− metastatic breast cancer: overview for oncology clinical practice. J. Mol. Pathol. 2, 42–54 (2021).
Zardavas, D. et al. Tumor PIK3CA genotype and prognosis in early-stage breast cancer: a pooled analysis of individual patient data. J. Clin. Oncol. 36, 981–990 (2018).
Luen, S. J. et al. Association of somatic driver alterations with prognosis in postmenopausal, hormone receptor-positive, HER2-negative early breast cancer: a secondary analysis of the BIG 1-98 randomized clinical trial. JAMA Oncol. 4, 1335–1343 (2018).
Mosele, F. et al. Outcome and molecular landscape of patients with PIK3CA-mutated metastatic breast cancer. Ann. Oncol. 31, 377–386 (2020).
Pang, B. et al. Prognostic role of PIK3CA mutations and their association with hormone receptor expression in breast cancer: a meta-analysis. Sci. Rep. 4 (2014).
Sobhani, N. et al. The prognostic value of PI3K mutational status in breast cancer: a meta-analysis. J. Cell. Biochem. 119, 4287–4292 (2018).
André, F. et al. Alpelisib for PIK3CA-mutated, hormone receptor–positive advanced breast cancer. N. Engl. J. Med. 380, 1929–1940 (2019).
Kadota, M. et al. Identification of novel gene amplifications in breast cancer and coexistence of gene amplification with an activating mutation of PIK3CA. Cancer Res. 69, 7357–7365 (2009).
Gonzalez-Angulo, A. M. et al. Frequency of mesenchymal-epithelial transition factor gene (MET) and the catalytic subunit of phosphoinositide-3-kinase (PIK3CA) copy number elevation and correlation with outcome in patients with early stage breast cancer. Cancer 119, 7–15 (2013).
Wu, G. et al. Somatic mutation and gain of copy number of PIK3CA in human breast cancer. Breast Cancer Res. 7, (2005).
Lee, M. H., Cho, J. H., Kwon, S. Y., Jung, S. J. & Lee, J. H. Clinicopathological characteristics of PIK3CA mutation and amplification in Korean patients with breast cancers. Int. J. Med. Sci. 17, 1131–1135 (2020).
Hosseini, S. et al. Relationship between PIK3CA amplification and P110α and CD34 tissue expression as angiogenesis markers in Iranian women with sporadic breast cancer. Iran. J. Pathol. 13, 447–453 (2018).
Campbell, I. G. et al. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 64, 7678–7681 (2004).
Firoozinia, M., Jahromi, M. Z., Moghadamtousi, S. Z., Nikzad, S. & Kadir, H. A. PIK3CA gene amplification and PI3K p110α protein expression in breast carcinoma. Int. J. Med. Sci. 11, 620–625 (2014).
Loibl, S. et al. Mutational diversity and therapy response in breast cancer: a sequencing analysis in the neoadjuvant geparsepto trial. Clin. Cancer Res. 25, 3986–3995 (2019).
López-Knowles, E. et al. PI3K pathway activation in breast cancer is associated with the basal-like phenotype and cancer-specific mortality. Int. J. Cancer 126, 1121–1131 (2010).
Fumagalli, D. et al. Somatic mutation, copy number and transcriptomic profiles of primary and matched metastatic estrogen receptor-positive breast cancers. Ann. Oncol. 27, 1860–1866 (2016).
Chia, S. K. L. et al. PIK3CA alterations and benefit with neratinib: analysis from the randomized, double-blind, placebo-controlled, phase III ExteNET trial. Breast Cancer Res. 21, 1–9 (2019).
Curtis, C. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012).
Pereira, B. et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun. 7, (2016).
Razavi, P. et al. The genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell 34, 427–438.e6 (2018).
Hoadley, K. A. et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell 173, 291–304.e6 (2018).
Yang, W. et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 41, D955–D961 (2013).
Gao, H. et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat. Med. 21, 1318–1325 (2015).
Razavi, P. et al. Alterations in PTEN and ESR1 promote clinical resistance to alpelisib plus aromatase inhibitors. Nat. Cancer 1, 382–393 (2020).
Nixon, M. J. et al. PIK3CA and MAP3K1 alterations imply luminal A status and are associated with clinical benefit from pan-PI3K inhibitor buparlisib and letrozole in ER+ metastatic breast cancer. npj Breast Cancer 5 (2019).
Mayer, I. A. et al. A phase Ib study of alpelisib (BYL719), a PI3Kα-specific inhibitor, with letrozole in ER+/HER2- metastatic breast cancer. Clin. Cancer Res. 23, 26–34 (2017).
Barbareschi, M. et al. Different prognostic roles of mutations in the helical and kinase domains of the PIK3CA gene in breast carcinomas. Clin. Cancer Res. 13, 6064–6069 (2007).
Vasan, N. et al. Double PIK3CA mutations in cis increase oncogenicity and sensitivity to PI3Ka inhibitors. Science 366, 714–723 (2019).
Zhang, Y. et al. A Pan-cancer proteogenomic atlas of PI3K/AKT/mTOR pathway alterations. Cancer Cell 31, 820–832.e3 (2017).
Smith, J. C. & Sheltzer, J. M. Systematic identification of mutations and copy number alterations associated with cancer patient prognosis. eLife 7 (2018).
Wilson, T. R. et al. The molecular landscape of high-risk early breast cancer: comprehensive biomarker analysis of a phase III adjuvant population. npj Breast Cancer 2, 1–9 (2016).
Boberg, D. R. et al. Copy number variation in ACHE/EPHB4 (7q22) and in BCHE/MME (3q26) genes in sporadic breast cancer. Chem. Biol. Interact. 203, 344–347 (2013). Chem Biol Interact.
Cerami, E. et al. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6 (2013).
Olshen, A. B., Venkatraman, E. S., Lucito, R. & Wigler, M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 5, 557–572 (2004).
Mermel, C. H. et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 12, R41 (2011).
Martínez-Jiménez, F. et al. A compendium of mutational cancer driver genes. Nat. Rev. Cancer 20, 555–572 (2020).
Acknowledgements
This study was supported by the Fondazione “Sandro Pitigliani” per la ricerca sul cancro ONLUS.
Author information
Authors and Affiliations
Contributions
I.M., M.P., E.R., L.M. and M.B. contributed to the conception and design of the study; I.M., M.P., C.B. and M.B. contributed to the acquisition and the analysis of the data; I.M., M.P., E.R., C.B., M.B. and L.M. contributed to the interpretation of the data., I.M. and M.P. drafted the manuscript; E.R., C.B., L.B., M.B. and L.M. substantially revised the manuscript; I.M., M.P., E.R., C.B., L.B., M.B. and L.M. approved the submitted version of the manuscript and have agreed both to be personally accountable for the author’s own contributions and to ensure that questions related to the accuracy or integrity of any part of the work, even ones in which the author was not personally involved, are appropriately investigated, resolved, and the resolution documented in the literature. I.M. and M.P. contributed equally. M.B. and L.M. jointly supervised this work.
Corresponding author
Ethics declarations
Competing interests
The authors declare the following competing interests: E.R.: Personal financial interests (honoraria, consulting or advisory role): Lilly, Roche, Pfizer, Amgen; L.B.: Personal financial interests (honoraria, consulting or advisory role): AstraZeneca, Daiichi-Sankyo, Eisai, Genomic Health, Lilly, Novartis, Pfizer, Pierre Fabre. Institutional financial interests: Celgene, Genomic Health, Novartis; M.B.: Consultant honoraria: Novartis; L.M.: Speaker/consultant honoraria: Novartis, Pfizer, Lilly. Research Grant: Novartis, Pfizer. The remaining authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Migliaccio, I., Paoli, M., Risi, E. et al. PIK3CA co-occurring mutations and copy-number gain in hormone receptor positive and HER2 negative breast cancer. npj Breast Cancer 8, 24 (2022). https://doi.org/10.1038/s41523-022-00382-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41523-022-00382-5