Abstract
Atrioventricular nodal reentry tachycardia (AVNRT) is the most common form of regular paroxysmal supraventricular tachycardia. This arrhythmia affects women twice as frequently as men, and is often diagnosed in patients <40 years of age. Familial clustering, early onset of symptoms and lack of structural anomaly indicate involvement of genetic factors in AVNRT pathophysiology. We hypothesized that AVNRT patients have a high prevalence of variants in genes that are highly expressed in the atrioventricular conduction axis of the heart and potentially involved in arrhythmic diseases. Next-generation sequencing of 67 genes was applied to the DNA profile of 298 AVNRT patients and 10 AVNRT family members using HaloPlex Target Enrichment System. In total, we identified 229 variants in 60 genes; 215 missenses, four frame shifts, four codon deletions, three missense and splice sites, two stop-gain variants, and one start-lost variant. Sixty-five of these were not present in the Exome Aggregation Consortium (ExAC) database. Furthermore, we report two AVNRT families with co-segregating variants. Seventy-five of 284 AVNRT patients (26.4%) and three family members to different AVNRT probands had one or more variants in genes affecting the sodium handling. Fifty-four out of 284 AVNRT patients (19.0%) had variants in genes affecting the calcium handling of the heart. We furthermore find a large proportion of variants in the HCN1-4 genes. We did not detect a significant enrichment of rare variants in the tested genes. This could be an indication that AVNRT might be an electrical arrhythmic disease with abnormal sodium and calcium handling.
Similar content being viewed by others
Introduction
Atrioventricular nodal reentry tachycardia (AVNRT) is a supraventricular tachycardia (SVT), originating from the atrioventricular node (AVN) region [1]. It is the most common form of regular paroxysmal SVT [2]. Women are affected twice as frequently as men, and the fact that this arrhythmia is often diagnosed in women under 40 years of age suggests a genetic component of the disease [3, 4]. The symptoms of SVT are typically related to the sudden occurrence of tachycardia, with palpitations being the most common symptom, possibly accompanied by chest discomfort, dyspnoea, anxiety, and light headedness. Some patients occasionally experience an unregulated drop in blood pressure at the onset of the tachycardia, resulting in syncope, particularly at high frequency rates and during prolonged episodes of tachycardia [5].
AVNRT occurs when a reentry circuit forms within or in close proximity to the AV node. Two areas with diverse electrophysiological conduction properties (termed pathways) have been found implicated in the reentry circuits in AVNRT patients, the fast pathway and the slow pathway; both located in the right atrium. These pathways show opposite electrophysiological properties, with respect to conduction velocity and duration of the refractory period, resulting in different relations between the P wave and the QRS complex on the electrocardiogram. In both cases, the P wave appearance is negative in leads II, III and aVF [6].
The presence of this diversity in the electrophysiological properties in the area around the AVN is termed dual AV nodal conduction. Dual AV nodal conduction is considered a congenital functional abnormality developed during cardiogenesis in foetal life [7]. In 2000, Lu et al. [8] described the occurrence of dual AV nodal conduction in monozygotic twins. In addition, Hayes et al. [7] described several families with AVNRT among first-degree relatives. The authors were also able to induce typical AVNRT and to demonstrate dual AV nodal conduction in 12 of the 13 studied family members from six different families [7]. A recent study by Michowitz et al. [9] reported a high familial AVNRT prevalence among patients who underwent radiofrequency ablation. These studies indicate that there could be a hereditary component in the development of AVNRT. Familial clustering, early onset of symptoms and the lack of structural anomaly indicate involvement of genetic factors, as seen in other arrhythmias [10]. Several cardiac arrhythmias have previously been associated with ion channel variants [11], and this may be a possible pathophysiological mechanism of AVNRT as well. Identifying possible disease-causing and disease-modifying genetic variants could potentially reveal new insight in the pathophysiology of AVNRT and have a role in future diagnosis and risk assessment. We hypothesized that AVNRT is an ion channel disease, and that AVNRT patients have a high prevalence of variants in genes, that are highly expressed in the atrioventricular conduction axis of the heart and are known to be involved in the pathophysiology of other arrhythmic diseases.
Subjects and Methods
Study population
Study participants (probands) were identified among patients treated with radiofrequency catheter ablation at the Department of Cardiology, Copenhagen University Hospital, Rigshospitalet, in the period from 2010 to 2012, with an age >18 years and <60 years. Furthermore, 10 relatives of these probands with a history of AVNRT were included. Upon inclusion, a blood sample, a 12 lead electrocardiogram (ECG), and a cardiac history were taken and patients were questioned about their family history.
The study conforms to the principles outlined in the Declaration of Helsinki, and was approved by the Scientific Ethics Committee of Copenhagen and Frederiksberg (protocol reference number: H-A-2008-004).
Control group
The control group consisted of 383 healthy men and women between 55 and 75 years of age and without history of cardiovascular disease or stroke from the Copenhagen Holter Study [12].
The study protocol was approved by the local ethics committee (KF 01 313322, KF 01 25304).
Target genes
We selected 67 target genes. The genes were selected based on the following criteria: (1) PR interval-associated genes identified by genome-wide association studies [13], (2) genes selected based on cardiac expression levels [14], (3) plausible genes based on protein function and association with other arrhythmic diseases. Selected genes are listed in Table 1.
Next-generation sequencing
DNA was extracted from whole blood that had been stored at −80 °C using the QIAamp DNA Blood Mini and Maxi kits (Qiagen, Hilden, Germany).
We developed a custom design based on HaloPlex technology (Agilent Technologies, Inc., Santa Clara, CA, USA) to perform high-throughput sequencing of the coding regions of 67 genes.
Next-generation sequencing (NGS) was applied using HaloPlex Target Enrichment System (Agilent Technologies, Inc.) on 200 ng DNA from the 298 probands and 10 family members with AVNRT according to the manufacturer’s instructions [15]. In brief, patient DNA was fragmented by endonucleases and hybridized to biotinylated gene specific probes incorporating Illumina paired-end sequencing motifs and indexed primers. Hybridized molecules were captured by magnetic beads, PCR amplified, and sequenced with the MiSeq system (Illumina Inc., San Diego, CA, USA).
Microarray genotyping
Genotyping was done using Infinium PsychArray BeadChip. This microarray has ~590,000 fixed markers. Single-nucleotide polymorphism calling and quality control (QC) and was done following Broad Institute’s recommendations.
In vitro electrophysiology
See Supplementary Material.
Bioinformatics and data analyses
Raw reads were aligned to reference genome GRCH37.p13/hg19 with Burrows-Wheelers Aligner after trimming adaptor sequences and filtering for poor quality reads. Genome Analysis Toolkit (GATK) v3.7 was used for indel realignment and base quality recalibration in the targeted regions. Variants were called with Unifiedgenotyper/GATK v3.7 following the GATK of Broad Institute’s current guidelines [16], see Fig. 1 for bioinformatics pipeline.
Diagram for the bioinformatics pipeline of sequence data. Data processing starts with raw reads from the next-generation sequencing. Reads, alignments, and variant calls are quality controlled (QC) in intermediate steps. Variants that are of interest for further analyses are obtained from a high-quality genotype set at the end of pipeline
For details on settings and filtering steps, see Supplementary Material.
Genotype array data were used to infer ethnicity and relatedness performed in R with R-package SNPRelate [17], see Supplementary Material.
Statistical analyses
We performed a burden test on the targeted genes with ethnically matched and unrelated subjects in the AVNRT cohort (considered subjects n = 284) and the control group (considered subjects n = 377). Only the intersecting region between the two capture kits and exons with sufficient coverage were considered in the tests, see Supplementary Figures 1, 2, 3 and Supplementary text. The intersect region spanned 30 genes, listed in Supplementary Table 7.
Variants with annotation of a putative deleterious impact were included, see Supplementary Table 3 (e.g., missense, codon deletions). Variants tagged as ‘COMMON’ in dbSnp b.141 were excluded.
We performed two rounds of burden testing. First, variants found in the Exome Aggregation Consortium (ExAC) database with a minor allele frequency (MAF) above 0.5% in any of the ExAC populations were excluded, second, this threshold was set to 0.1%. The statistical method SKAT-O was applied in the burden tests, using R package GENESIS [18, 19]. A Bonferroni-corrected p value < 0.05 was considered significant.
Data availability
Data on reported variants are made available to the European Bioinformatics Institute (EBI) database The European Genome-phenome Archive (EGA) (https://www.ebi.ac.uk/ega/, study accession EGAS00001002745).
Results
Two-hundred-and-eighty-four patients with a median onset of AVNRT-related symptoms at 24 years of age were included after excluding ethnic outliers and subjects with unreported relatedness (70% females). Clinical data of the patient population (probands) are shown in Table 2.
We identified 229 variants in 184 patients; 215 missense, four frame shift, four codon deletions, three missense and splice sites, two stop-gain variants, and one start-lost variant. Sixty-five of these were not present in ExAC. The results of the NGS are shown in Supplementary Table 4.
Variants in sodium-handling genes
There were 75 out of 284 AVNRT patients (26.4%) and three family members of three different AVNRT probands who had one or more variants in genes affecting the sodium handling.
We identified variants in SCN3A (n = 3, ENSG00000166257.4), SCN5A (n = 7, ENSG00000153253.11), SCN10A (n = 16, ENSG00000185313.6), SCN8A (n = 3, ENSG00000196876.9), SCN4A (n = 12, ENSG00000007314.7), SCN1A (n = 3, ENSG00000144285.11), SCN2B (n = 1, ENSG00000149575.5) and SCN9A (n = 12, ENSG00000169432.10).
Variants in calcium-handling genes
Fifty-four of 284 AVNRT patients (19.0%) had variants in genes affecting the calcium handling of the heart. We identified variants in RYR2 (n = 11, ENSG00000198626.11), RYR3 (n = 16, ENSG00000198838.7), CACNB2 (n = 2, ENSG00000165995.14), ATP2A2 (n = 2, ENSG00000174437.12), CACNA1C (n = 5, ENSG00000151067.16), CACNA1D (n = 13, ENSG00000157388.9), CACNA1I (n = 7, ENSG00000100346.13) and CACNA1G (n = 3, ENSG00000006283.13).
Variants in the HCN1-4 genes
Thirteen AVNRT patients carried variants in the hyperpolarization-activated and cyclic nucleotide-gated (HCN) channel genes HCN1 (n = 5, ENSG00000164588.4), HCN2 (n = 1, ENSG00000099822.2), HCN3 (n = 5, ENSG00000263324.1) and HCN4 (n = 2, ENSG00000138622.3).
Variants in KCNE3
Three variants with a total allele count of four were found in the KCNE3 (ENSG00000175538.6) gene encoding the voltage-gated potassium channel Kv.
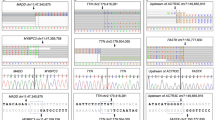
Family studies
Six families were identified with AVNRT reported in two or more family members. In two of these families, variants within the 67 screened genes where found (Fig. 2). The proband carrying the c.6010T>C (p.(Phe2004Leu), ENST00000413689) variant in SCN5A and the c.2623C>T (p.(Pro875Ser), ENST00000435607) variant in SCN4A had a mother with AVNRT who carried the SCN4A variant and a sister with AVNRT who carried both the SCN5A and the SCN4A variant. Furthermore, the mother and the sister with AVNRT both carried the RYR3 variant c.3598C>T (p.(Arg1200Cys), ENST00000389232).
The proband carrying the c.233C>T (p.(Thr78Met, ENST00000343849)) variant in CAV3 had a mother with AVNRT who carried the same variant. For pedigrees of the four AVNRT families without found variants in the 67 screened genes, see Supplementary Figure 6.
Electrophysiological characterization of the SCN5A variants c.1381C>T and c.1576C>T
As numerous genetic variations in SCN5A (ENSG00000183873.11), encoding the primary cardiac sodium channel Nav1.5, have been linked to a number of arrhythmogenic diseases, it is likely that malfunction of Nav1.5 may also play a role in AVNRT. A total of seven variants in SCN5A were found in the 284 AVNRT patients. Four of these have previously been functionally characterized by patch-clamping and the fifth has a stop-gain translation impact, indicating a compromised sodium channel function [20, 21].
The in vitro electrophysiological characteristics of c.1381C>T (p.(Leu461Val)) and c.1576C>T (p.(Arg526Cys)) were studied by patch-clamping following transient expression in HEK293 cells (Fig. 3). Whole-cell sodium currents from wildtype (WT) and variant channels in response to depolarizing pulses from −70 to +50 mV are shown in Fig. 3a. Peak current density was significantly increased for c.1381C>T (p.(Leu461Val)) but not for c.1576C>T (p.(Arg526Cys)) (Supplementary Table 5). Activation and inactivation together with time-dependent inactivation (onset of inactivation) and recovery from inactivation, as well as sustained current measurements were performed. However, none of these investigations revealed a difference between WT and variant channels.
Electrophysiological effects of Nav1.5-WT, L461V and R526C on sodium channel current (INa). Whole-cell patch clamp analyses of transiently transfected HEK-293 cells. a Representative INa traces in cells expressing WT or mutants. b Peak current–voltage relationship measured at −20 mV for Nav1.5-WT (n = 7), L461V (n = 10) and R526C (n = 7). Currents were normalized to membrane capacitance. R526C was significantly different from WT (*p < 0.05). c, d Voltage dependence of activation and inactivation for Nav1.5 of the three groups, indicating gating properties of channel conductance and availability. The normalized values have been calculated by dividing the current level at the respective voltage by the maximal current of the whole-voltage range (I/Imax). Boltzmann curves were fitted to both steady-state activation and inactivation data. Averaged values and the number of cells used are represented in Supplementary Table 5. e Time dependent of recovery from fast inactivation of three groups determined using a two-pulse protocol. Data were fitted with a single exponential equation. Time constants are listed in Supplementary Table 5. The applied voltage-clamp protocols in inset of the respective figure. Of note, a recovery potential of −80 mV was applied in order to mimic the resting membrane potential in the human atrial myocytes. f Sustained (late) Na+ current as percentage of peak current measured following 330–350 ms depolarizations to −20 mV. g Onset (decay) of inactivation following depolarizing pulses to either −20, 0 or 20 mV. c and d were analysed by Student’s unpaired t test. e, f and g were analysed by one-way ANOVA repeated measures with Dunnett’s post-test
Burden tests
After exclusion of family members and ethnic outliers, the data set consisted of 284 AVNRT cases versus 377 controls.
There was no significant enrichment of rare variants in the tested genes after adjusting for multiple testing, see Supplementary Table 6 and 7.
Discussion
In total, we identified 229 variants in 184 patients; 215 missense, four frame shift, four codon deletions, three missense and splice sites, two stop-gain variants, and one start-lost variant. Sixty-five of these were not present in ExAC.
A recent study by Hasdemir et al. [22] found a phenotypic overlap between 17 patients with AVNRT and Brugada Syndrome, and following genetic screening recognized a high proportion of sodium channel variation in this cohort. Their findings indicate a possible sodium channel abnormality in AVNRT patients.
Here we give a descriptive study that characterize the genetic component of AVNRT, with a focus on variation in SCN5A as affected conduction velocity was considered a possible disease mechanism.
A total of seven variants in SCN5A were found in the 284 AVNRT patients. Four of these have previously been functionally characterized by patch-clamping and the fifth has a stop-gain translation impact, indicating a compromised sodium channel function [20, 21]. In a study on sudden infant death syndrome (SIDS), Wang et al. [21] described the c.6010T>C (p.(Phe2004Leu)) variant in SCN5A to have a gain-of-function effect. The c.1019G>A (p.(Arg340Gln)) variant in SCN5A has been associated with SIDS [21] and long QT syndrome 3 and has been shown to induce a negative voltage shift of both steady-state activation and inactivation together with a reduced time constant for onset of fast inactivation [20]. We performed functional studies on two of the remaining variants. Our functional studies of c.1381C>T (p.(Leu461Val)) and c.1576C>T (p.(Arg526Cys)) in SCN5A (ENST00000413689) revealed an increased current density for c.1576C>T, while c.1381C>T was not found to have altered current characteristics on the parameters tested. Our subsequent principal component analysis indicated that the carrier had a mixed ethnic background (Supplementary Figure 5a) and further analyses of the variant showed a MAF >1% within an Afro-American cohort [23]. We identified 16 variants in the SCN10A gene, whereof four had been functionally characterized in previous studies and the fifth has a stop-gain translation impact [24, 25].
Although functional characterization not equals causality, our results might indicate that altered sodium current predispose for AVNRT by affecting the conduction velocity due to potential disease modifiers in sodium-handling genes. Independent replication of these results is, however, needed in larger cohorts.
It is also noteworthy that we identified two loss-of-function (LOF) variants in the KCNE3 gene, which has only three reported LOF variants in the ExAC database.
It is difficult to conclude on these findings, but nevertheless interesting.
Several studies have found AVNRT in atrial fibrillation (AF) patients, and this overlap in arrhythmia phenotype may support an overlap in disease mechanisms [25,26,27]. This suggested common disease mechanism is supported by the fact that several studies have demonstrated frequent sodium channel variants in AF patients. The results from the present study combined with the data by Jabbari et al. [25] who sequenced 225 early-onset lone AF patients and found 11 patients with rare variants in SCN10A of which three (27%) had an AVNRT diagnosis as well, despite all of them being diagnosed with AF before the age of 36, further supports the involvement of sodium channels in the pathophysiology of AVNRT.
Some of our AVNRT probands carried variants in the HCN1-4 genes, which have all been found to have a high intolerance to variation (a positive Z-score up to 7.27 for HCN2 in the ExAC browser [23]). The HCN2 and HCN4 genes have previously been associated with sinoatrial nodal dysfunction and the HCN genes have an important role in the contractility of the heart muscle by restoring the resting membrane potential in cardiomyocytes from hyperpolarized potentials as well as contributing to the next depolarization [28].
Interestingly, the c.2623C>T (p.(Pro875Ser), ENST00000435607) variant in SCN4A co-segregated in a family with three AVNRT patients, a mother and her two daughters, see Fig. 2. Traditionally, the SCN4A gene is not considered a main contributor to the electrophysiology of the heart, but our findings, although only hypothesis generating, might indicate a role for Nav1.4 in the atria.
One of the challenges with AVNRT is that its definite diagnosis requires invasive electrophysiological study (EPS) or an oesophageal ECG recording. To obtain the highest degree of correct clinical phenotype in this study, we required that patients had an invasive EPS demonstrating AVNRT and following this the patients had a radiofrequency catheter ablation performed. By these relatively strong inclusion criteria, we have minimized the risk of misdiagnosis of tachyarrhythmia subtype. It is important to consider that studies have found an overrepresentation of previously phenotype-associated genetic variants in the general population [29, 30]. This indicates that some of these genetic variants might not be monogenic causes of disease. Also, the functional analyses performed in this study used a conventional heterologous expression system; hence, the environments differ from that in native cardiomyocytes. Lastly, as dual AV nodal physiology can be asymptomatic, it would be interesting to test control subjects for dual AV nodal physiology in a larger study setup.
This is, to our knowledge, the first study where the primary aim was to investigate the genetic component in AVNRT using a NGS approach. We have given a descriptive report of variants seen in genes responsible for the sodium of the heart, many of which have been functionally characterized. Furthermore, we report two AVNRT families with co-segregating variants. Despite of familial clustering, we are not able to detect a single gene or gene family to be involved with AVNRT, however, we do report several interesting findings of rare genetic variation in AVNRT. The data suggest that some patients with AVNRT have genetic variants that can potentially affect sodium handling, possibly by affecting the conduction velocity and the refractory period. The genetic picture is, however, still complex, and structural genes might play a larger role in cardiac arrhythmias than previously thought. As this is only the beginning of the genetic investigation of AVNRT, the need for further genetic studies is needed.
References
Katritsis DG, Camm AJ. Classification and differential diagnosis of atrioventricular nodal re-entrant tachycardia. Europace. 2006;8:29–36.
Katritsis DG, Camm AJ. Atrioventricular nodal reentrant tachycardia. Circulation. 2010;122:831–40.
Kwaku KF, Josephson ME. Typical AVNRT--an update on mechanisms and therapy. Card Electrophysiol Rev. 2002;6:414–21.
Marchlinski FE. Diagnosing the mechanism of supraventricular tachycardia: restoring the luster of a fading art. J Am Coll Cardiol. 2009;53:2359–61.
Medi C, Kalman JM, Freedman SB. Supraventricular tachycardia. Med J Aust. 2009;190:255–60.
Akhtar M, Jazayeri MR, Sra J, Blanck Z, Deshpande S, Dhala A. Atrioventricular nodal reentry (AVNRT) clinical, electrophysiological, and therapeutic considerations. Circulation. 1993;88:282–95.
Hayes JJ, Sharma PP, Smith PN, Vidaillet HJ. Familial atrioventricular nodal reentry tachycardia. Pacing Clin Electrophysiol. 2004;27:73–6.
Lu CW, Wu MH, Chu SH. Paroxysmal supraventricular tachycardia in identical twins with the same left lateral accessory pathways and innocent dual atrioventricular pathways. Pacing Clin Electrophysiol. 2000;23:1564–6.
Michowitz Y, Anis-Heusler A, Reinstein E, Tovia-Brodie O, Glick A, Belhassen B. Familial occurrence of atrioventricular nodal reentrant tachycardia. Circ Arrhythm Electrophysiol. 2017;10:e004680.
Øyen N, Ranthe MF, Carstensen L, et al. Familial aggregation of lone atrial fibrillation in young persons. J Am Coll Cardiol. 2012;60:917–21.
Priori SG, Blomström-Lundqvist C, Mazzanti A. et al. 2015 ESC Guidelines for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac death: the task force for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac death of the European Society of Cardiology (ESC). Endorsed by: Association for European Paediatric and Congenital Cardiology (AEPC). Eur Heart J. 2015;36:2793–867.
Sajadieh A, Nielsen OW, Rasmussen V, Hein HO, Hansen JF. Prevalence and prognostic significance of daily-life silent myocardial ischaemia in middle-aged and elderly subjects with no apparent heart disease. Eur Heart J. 2005;26:1402–9.
Pfeufer A, van Noord C, Marciante KD, et al. Genome-wide association study of PR interval. Nat Genet. 2010;42:153–9.
Greener ID, Monfredi O, Inada S, et al. Molecular architecture of the human specialised atrioventricular conduction axis. J Mol Cell Cardiol. 2011;50:642–51.
Agilent. HaloPlex Target Enrichment System http://www.chem.agilent.com/library/usermanuals/Public/G9900-90001.pdf (2014).
DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8.
Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS. A high-performance computing toolset for relatedness and principal component analysis of SNPdata. Bioinforma Oxf Engl. 2012;28:3326–8.
Ionita-Laza I, Lee S, Makarov V, Buxbaum JD, Lin X. Sequence kernel association tests for the combined effect of rare and common variants. Am J Hum Genet. 2013;92:841–53.
Bioconductor—GENESIS (development version) https://www.bioconductor.org/packages/devel/bioc/html/GENESIS.html (2017).
Olesen MS, Yuan L, Liang B, et al. High prevalence of long QT syndrome-associated SCN5A variants in patients with early-onset lone atrial fibrillation. Circ Cardiovasc Genet. 2012;5:450–9.
Wang DW, Desai RR, Crotti L, et al. Cardiac sodium channel dysfunction in sudden infant death syndrome. Circulation. 2007;115:368–76.
Hasdemir C, Payzin S, Kocabas U, et al. High prevalence of concealed Brugada syndrome in patients with atrioventricular nodal reentrant tachycardia. Heart Rhythm. 2015;12:1584–94.
ExAC Browser http://exac.broadinstitute.org/ (2015).
Savio-Galimberti E, Weeke P, Muhammad R, et al. SCN10A/Nav1.8 modulation of peak and late sodium currents in patients with early onset atrial fibrillation. Cardiovasc Res. 2014;104:355–63.
Jabbari J, Olesen MS, Yuan L, et al. Common and rare variants in SCN10A modulate the risk of atrial fibrillation. Circ Cardiovasc Genet. 2015;8:64–73.
Palma EC, Ferrick KJ, Gross JN, Kim SG, Fisher JD. Transition from atrioventricular node reentry tachycardia to atrial fibrillation begins in the pulmonary veins. Circulation. 2000;102:937.
Brugada J, Mont L, Matas M, Navarro-López F. Atrial fibrillation induced by atrioventricular nodal reentrant tachycardia. Am J Cardiol. 1997;79:681–2.
Günther A, Baumann A. Distinct expression patterns of HCN channels in HL-1 cardiomyocytes. BMC Cell Biol. 2015;16:18.
Andreasen C, Nielsen JB, Refsgaard L, et al. New population-based exome data are questioning the pathogenicity of previously cardiomyopathy-associated genetic variants. Eur J Hum Genet. 2013;21:918–28.
Ghouse J, Have CT, Weeke P, et al. Rare genetic variants previously associated with congenital forms of long QT syndrome have little or no effect on the QT interval. Eur Heart J. 2015;36:2523–9.
Acknowledgements
This work was supported by a framework grant of the Danish National Research Foundation, the Danish Council for Independent Research (DFF-1331-00313B), The Lundbeck Foundation, The Villadsen Family Foundation, The John and Birthe Meyer Foundation, The Arvid Nilsson Foundation, Direktør Ib Henriksens Fond and Fondsbørsvekselerer Henry Hansen og Hustru Karla Hansen, født Westergaards, legat, and The Research Council at Rigshospitalet. We would like to thank Katrine Kastberg for laboratory work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Laura Andreasen, Gustav Ahlberg, Morten S. Olesen and Jesper H. Svendsen contributed equally to this work.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Andreasen, L., Ahlberg, G., Tang, C. et al. Next-generation sequencing of AV nodal reentrant tachycardia patients identifies broad spectrum of variants in ion channel genes. Eur J Hum Genet 26, 660–668 (2018). https://doi.org/10.1038/s41431-017-0092-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-017-0092-0