Abstract
As a core kinase of antiviral immunity, the activity and stability of TANK-binding kinase 1 (TBK1) is tightly controlled by multiple post-translational modifications. Although it has been demonstrated that TBK1 stability can be regulated by ubiquitin-dependent proteasome pathway, it is unclear whether another important protein degradation pathway, autophagosome pathway, can specifically affect TBK1 degradation by cargo receptors. Here we report that E3 ubiquitin ligase NEDD4 functions as a negative regulator of type I interferon (IFN) signaling by targeting TBK1 for degradation at the late stage of viral infection, to prevent the host from excessive immune response. Mechanically NEDD4 catalyzes the K27-linked poly-ubiquitination of TBK1 at K344, which serves as a recognition signal for cargo receptor NDP52-mediated selective autophagic degradation. Taken together, our study reveals the regulatory role of NEDD4 in balancing TBK1-centered type I IFN activation and provides insights into the crosstalk between selective autophagy and antiviral signaling.
Similar content being viewed by others
Introduction
The innate immune system, an evolutionarily conserved defense strategy, provides the first line of host defense against invading microbial pathogens [1]. During viral infection, the pattern recognition receptors (PRRs) of host cells, such as Toll-like receptors (TLRs), RIG-I-like receptors (RLRs), NOD-like receptors (NLRs) and several nucleic acid sensors, recognize viral pathogen-associated molecular patterns (PAMPs) and initiate antiviral responses by triggering the production of proinflammatory cytokines and type I interferons (IFNs) [2, 3]. Subsequently, IFN eliminates the invading viruses and provokes the comprehensive antiviral states via induction of hundreds of IFN-stimulated genes (ISGs) [4].
TBK1 is a key kinase for IFN production and plays important role in antiviral innate immune responses. Upon viral infection, TBK1 triggers phosphorylation and translocation of IFN-regulatory factor 3 (IRF3), which then promotes transcriptional activation of type I IFNs [5]. TBK1 activity is tightly regulated by multiple post-translational modifications, such as phosphorylation, ubiquitination, sumoylation, and acetylation [6,7,8]. Autophosphorylation of TBK1 at Ser (S) 172, induced by glycogen synthase kinase 3β (GSK3β), is indispensable for its activation [9]. In addition, the stability of TBK1 is also essential for its function to modulate type I IFN signaling. TBK1 can be degraded through ubiquitin-proteasome pathway by many regulators, including NLRP4, DTX4, TRIP, TRIM27, USP38, NLRP14 and TRAF3IP3 [10,11,12,13,14,15]. However, whether TBK1 stability can be controlled by other degradation pathway during viral infection remains unknown.
Autophagy is a highly conserved intracellular degradation pathway that decomposes cytoplasmic components, organelles and invading pathogens, and plays important role in the cellular homeostasis, nutrient recycling and stress response [16, 17]. Accumulating evidence showed that autophagy is widely implicated with innate immunity [18]. On one hand, the innate immune system activates autophagy to degrade and dispose of invading pathogens [2]. On the other hand, many components of autophagy machinery can directly regulate innate immune signaling pathways, such as nuclear factor κB (NF-κB) and type I IFN signaling [3, 19]. Interestingly, TBK1 has also been reported to be involved in the regulation of autophagy. TBK1 phosphorylates syntaxin 17, which functions in assembly of protein complexes during autophagy initiation, leading to the formation of FIP200-ULK1-ATG13 complex [20]. Moreover, TBK1 promotes selective autophagy by phosphorylating cargo receptors, including OPTN, p62, NDP52 and TAX1BP1 [21]. These finding suggests that TBK1 might play important role in the crosstalk between antiviral immunity and autophagy. Recently, we and other researchers have demonstrated that selective autophagy mediated by diverse cargo receptors can subtly control the activation of innate immune signaling by degrading several major PRRs, adaptors, and transcriptional factors, including RIG-I, cGAS, AIM2, MAVS, STING, TRIF, and IRF3 [22,23,24,25,26,27,28,29]. However, whether kinases in type I IFN signaling are degraded through the selective autophagic pathway remains unclear.
Neural precursor cell expressed developmentally downregulated protein 4 (NEDD4, also known as NEDD4-1) belongs to the HECT ubiquitin E3 ligase family, and has been reported to play pivotal role in innate immunity and multifarious cellular processes, including cell proliferation, apoptosis and autophagy [30,31,32,33]. Several reports showed that NEDD4 positively regulates autophagy by modulating the ubiquitination of Beclin-1, VPS34 and p62 respectively [33,34,35]. Furthermore, NEDD4 contributes to the budding process of various viruses through ubiquitinating viral matrix proteins [34]. The ubiquitination activity of NEDD4 could be inhibited by ISG15 to enhance the innate antiviral response [36, 37], implying the critical role of NEDD4 in antiviral pathway. However, whether NEDD4 affects the crosstalk between antiviral immunity and autophagy remains unclear. Here, we investigated the mechanism by which NEDD4 negatively regulates type I IFN signaling to mediate the homeostasis of innate immunity. Upon viral infection, NEDD4 binds to the active form of TBK1 and catalyzes its K27-linked poly-ubiquitination via its E3 ligase activity. K27-linked poly-ubiquitin chains on TBK1 at Lys (K) 344 are recognized by the cargo receptor NDP52 and result in the selective autophagic degradation of TBK1. Our findings demonstrate that NEDD4 functions as a “molecular brake” to balance type I IFN response by targeting TBK1 through selective autophagy.
Results
NEDD4 negatively regulates type I IFN signaling
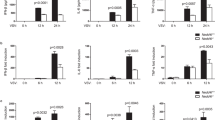
Mounting evidence revealed that HECT-type E3 ligase NEDD4 affects virus infection and budding [32, 34], yet the regulatory mechanisms remain poorly understood. Thus, we assessed the effects of NEDD4 on type I IFN signaling as well as antiviral immunity. We found that ectopic expression of NEDD4 substantially inhibited the activation of IFN-stimulated response element (ISRE) and IFN-β by infection of Sendai virus (SeV), vesicular stomatitis virus (VSV), or intracellular (IC) poly (I:C) treatment (Fig. 1a, b and Supplementary Fig. 1a, b). To determine the role of NEDD4 under physiological conditions, we efficiently knocked down NEDD4 in 293 T cells (Supplementary Fig. 1c) and found that NEDD4 depletion could enhance SeV, VSV, or IC poly (I:C)-induced ISRE activation (Fig. 1c and Supplementary Fig. 1d). To further demonstrate the role of NEDD4 in type I IFN signaling, we generated a Flag-tagged NEDD4 doxycycline (Dox)-inducible A549 cell line (Supplementary Fig. 1e), and found that ectopic expression of NEDD4 considerably inhibited IRF3 phosphorylation after SeV and VSV infection (Fig. 1d and Supplementary Fig. 1f). In contrast, knockout of NEDD4 remarkably increased the phosphorylation of IRF3 (Fig. 1e). In addition, we found that the mRNA levels of IFNB, ISG54 and ISG56 in NEDD4 overexpression cells were significantly reduced during SeV and VSV infection (Fig. 1f and Supplementary Fig. 1g, h, i). We next observed that ectopic expression of NEDD4 rendered the cells more susceptible to viral infection and resulted in considerably more GFP+ (virus-infected) cells than mock cells (Fig. 1g, h and Supplementary Fig. 1j). Collectively, these results suggest that NEDD4 negatively regulates type I IFN signaling as well as antiviral immunity.
Luciferase activity in 293 T cells transfected with an ISRE (a) or IFN-β (b) promoter-driven luciferase reporter, together with the indicated plasmids along with empty vector (no wedge) or increasing amounts (wedge) of expression vector for HA-NEDD4, followed by treatment with or without SeV (MOI = 0.1), VSV-eGFP (MOI = 0.1) or intracellular (IC) poly (I:C) (5 μg/mL) infection for 12 h respectively. c Luciferase activity in 293 T cells transfected with scrambled (Scr) siRNA or NEDD4-specific siRNAs, together with an ISRE-luc, then treated or untreated with SeV, VSV-eGFP or IC poly(I:C). d Flag-tagged NEDD4-inducible A549 cells were treated with Dox (100 ng/mL) for 12 h, followed by SeV (MOI = 0.1) infection at indicated time points. The lysate was harvested for immunoblot analysis with indicated antibodies. e Protein lysates of wild-type (WT) and NEDD4 knockout (KO) A549 cells infected with SeV (MOI = 0.1) at indicated time points were immunoblotted with indicated antibodies. f RT–PCR analysis of indicated gene expression in Flag-tagged NEDD4-inducible A549 cells treated with Dox (100 ng/mL) for 12 h, followed by SeV (MOI = 0.1) infection at indicated time points. Phase-contrast (PH) and fluorescence microscopy analyses (g) or flow cytometric analyses (h) of Flag-tagged NEDD4-inducible A549 cells treated with or without Dox (100 ng/mL) for 12 h, followed by VSV-eGFP (MOI = 0.01) infection at indicated time points. Scale bars, 200 μm. In (a–c and f), all error bars, mean values ± SEM, P values were determined by unpaired two-tailed Student’s t test of n = 3 independent biological experiments, **P < 0.01 and *** P < 0.001. For (d, e, g, and h), similar results are obtained for three independent biological experiments.
Effect of NEDD4 on the expression of type I IFN-induced ISGs by viral infection
We further performed global RNA-sequencing (RNA-seq) analysis to identify cells and pathways regulated by NEDD4. Differentially expression analysis identified 4216 up-regulated genes and 221 down-regulated genes in response to NEDD4 deficiency in A549 cells after SeV infection (Fig. 2a, b). Gene ontology (GO) analysis further demonstrated that these genes are involved in cellular immune responses, including “response to virus” (Fig. 2c). The abolishment of NEDD4 led to the up-regulation of a variety of ISGs (Fig. 2d). Moreover, Gene-set-enrichment analyses (GSEA) was performed to analyze IFN activity-related genes and demonstrated the core-enriched genes in antiviral immunity group, such as “interferon” (Fig. 2e). Differentially expression analysis further showed that NEDD4 deficiency results in the up-regulation of many type I IFN-induced ISGs after SeV infection (Fig. 2f). These results indicate that NEDD4 negatively regulates antivirus responses during viral infection by controlling the expression of type I IFNs and ISGs.
a Heatmap view of top and bottom gene list of RNA-sequence data sets. Microarray analysis for total RNA was performed for wild-type (WT) and NEDD4 knockout (KO) A549 cells with or without SeV infection. b NEDD4 regulates antivirus response-relevant target genes, presented as a volcano plot of genes with differential expression after SeV infection in WT and NEDD4 KO A549 cells. c Gene ontology enrichment analysis of the NEDD4-dependent genes in B (−log2 p Values). d The log2 Fold Change (FC) of indicated ISGs in WT and NEDD4 KO A549 cells with SeV infection. e GSEA of differentially expressed genes in NEDD4 KO A549 cells with SeV infection and enrichment of interferon. FDR (q-value) was shown. f Heatmap view of mRNA variations of type I IFN-mediated ISG sets in WT and NEDD4 KO A549 cells with or without SeV infection.
NEDD4 interacts with TBK1
To investigate the molecular target of NEDD4 in type I IFN signaling, we performed coimmunoprecipitation assay and found that NEDD4 interacted with TBK1 and IKKi, but not cGAS, STING, RIG-I, MDA5, MAVS, or IRF3 (Fig. 3a). In addition, luciferase reporter assay showed that NEDD4 inhibited the activation of luciferase reporters induced by RIG-I, MDA5, MAVS and TBK1, but not IKKi or IRF3 (5D) (a constitutively active mutant of IRF3) (Fig. 3b, c and Supplementary Fig. 2a, b), suggesting that NEDD4 inhibits type I IFN signaling at TBK1 level. We next performed mass spectrometry (MS) analysis of TBK1-associated proteins from 293 T cells with SeV infection, using a similar strategy as previously described [26], and MS analysis showed that TBK1 could interact with NEDD4 (Fig. 3d and Supplementary Fig. 2c). In addition, we found that the endogenous association between NEDD4 and TBK1 was markedly enhanced upon SeV and VSV infection (Fig. 3e and Supplementary Fig. 2d). This result is confirmed in PBMCs infected with SeV (Fig. 3f). Further confocal microscopy analysis revealed that some of NEDD4 and TBK1 expressed at the same location in uninfected cells and the colocalization between NEDD4 and TBK1 was increased upon SeV infection (Fig. 3g, h). Moreover, we found NEDD4 interacted with TBK1 through its HECT domain by coimmunoprecipitation assay (Fig. 3i). Collectively, these results suggest that NEDD4 suppresses type I IFN signaling by targeting TBK1.
a Coimmunoprecipitation and immunoblot analysis of 293 T cells transfected with HA-NEDD4 together with Flag-tagged cGAS, STING, RIG-I, MDA5, MAVS, TBK1, IKKi or IRF3. b, c 293 T cells were transfected with the ISRE (b) or IFN-β (c) luciferase reporter, together with the indicated plasmids along with empty vector (no wedge) or increasing amounts (wedge) of expression vector for HA-NEDD4. NS: not significant. d 293 T cells were transfected with Flag-TBK1 and followed by SeV (MOI = 0.1) infection for 12 h. Flag-tagged TBK1 purified from 293 T cells was resolved on SDS-PAGE and stained with Coomassie Blue. TBK1-binding peptides were identified in Flag-TBK1 by mass spectrometry analysis. Protein lysates of A549 cells (e) or PBMCs (f) infected with SeV (MOI = 0.1) for indicated time points (above lanes) were subjected to immunoprecipitation with anti-NEDD4 antibody and immunoblot analysis with indicated antibodies. g Confocal microscopy of A549 cells infected with SeV (MOI = 0.1) for 12 h before harvested, followed by labeling of NEDD4 and TBK1 with specific primary antibody and Alexa Fluor 488 goat anti-Rabbit IgG (H + L) (green) or CF568 donkey anti-goat-IgG secondary antibody (red). Scale bars, 20 μm. h Quantitative analysis of the colocalization (30 cells per group) in (g). i Coimmunoprecipitation and immunoblot analysis of 293 T cells transfected with vectors for NEDD4 and its deletions along with vector encoding Flag-TBK1. In (b, c), all error bars, mean values ± SEM, P values were determined by unpaired two-tailed Student’s t test of n = 3 independent biological experiments, **P < 0.01. In (h), all error bars, mean values ± SD of 30 cells. P values were determined by unpaired two-tailed Student’s t test, ***P < 0.001. For (a, e, f, g, and i), similar results are obtained for three independent biological experiments.
NEDD4 promotes the autophagic degradation of TBK1
We next sought to dissect the inhibitory function of NEDD4 in type I IFN signaling through its interaction with TBK1. In 293 T cells transfected with Flag-tagged TBK1 or IKKi, together with increasing doses of NEDD4, we found that increasing amount of NEDD4 considerably decreased the protein level of TBK1, but not IKKi (Fig. 4a and Supplementary Fig. 3a). However, the abundance of TBK1 mRNA remained unchanged (Fig. 4a), suggesting that NEDD4 promotes TBK1 protein degradation. To determine whether NEDD4 mediates the degradation of endogenous TBK1 under virus-infected conditions, we ectopically expressed NEDD4 and found that NEDD4 overexpression decreased the TBK1 protein abundance in SeV- and VSV-infected cells, but not in uninfected cells (Fig. 4b and Supplementary Fig. 3b). Furthermore, knockout of NEDD4 resulted in higher TBK1 protein levels in SeV-infected cells (Fig. 4c). To further confirm our findings, we knocked down NEDD4 in PBMCs and found that NEDD4 deficiency resulted in increased TBK1 abundance in SeV-infected human primary cells (Fig. 4d). To investigate whether the ubiquitin E3 ligase activity of NEDD4 is required to promote TBK1 degradation, we generated the construct encoding catalytically inactive mutant of NEDD4, NEDD4 HA/CA (H1284A/C1286A) and observed that NEDD4 mediated the degradation of TBK1 in a catalytic activity-dependent manner (Fig. 4e). Above results indicate that NEDD4 might specifically target the active form of TBK1 for degradation. Therefore, we co-transfected 293 T cells with NEDD4, wild type (WT) TBK1, or its mutant (S172A, which abrogates auto-phosphorylation of TBK1), and we found that NEDD4 could not interact with TBK1 S172A mutant and failed to promote its degradation (Fig. 4f, g). These results suggest that NEDD4 specifically promotes the degradation of activated TBK1 (p-TBK1) after viral infection.
a 293 T cells were transfected with Flag-TBK1, together with increasing amounts (wedge) of expression vector for HA-NEDD4, and the protein was harvested for immunoblot analysis. Below, RT-PCR analysis of TBK1 mRNA; RPL13 mRNA serves as a loading control. b Flag-tagged NEDD4-inducible A549 cells were treated with doxycycline (Dox) (100 ng/mL) for 12 h, followed by SeV (MOI = 0.1) infection at indicated time points. The lysate was harvested for immunoblot analysis with indicated antibodies. c Protein lysates of wild-type (WT) and NEDD4 knockout (KO) A549 cells infected with SeV (MOI = 0.1) at indicated time points were immunoblotted with indicated antibodies. d PBMCs were transfected with scrambled (Scr) siRNA or NEDD4-specific siRNAs, followed by SeV (MOI = 0.1) infection for 12 h, the lysates were then analyzed with indicated antibodies. e Immunoblot analysis of protein lysates of 293 T cells transfected with vectors expressing WT HA-NEDD4 or HA-NEDD4 HA/CA mutant, together with plasmids encoding Flag-TBK1. f Coimmunoprecipitation and immunoblot analysis of protein lysates of 293 T cells transfected with vectors expressing WT Flag-TBK1 or Flag-TBK1 S172A mutant, together with HA-NEDD4. g Immunoblot analysis of protein lysates of 293 T cells transfected with vectors expressing WT Flag-TBK1 or Flag-TBK1 S172A mutant, together with plasmids encoding empty vector (EV) or HA-NEDD4. h 293 T cells were transfected with plasmid encoding Flag-TBK1, together with plasmids encoding EV or HA-NEDD4, followed by treatments of MG132 (10 μM), 3MA (10 mM), CQ (50 μM), bafilomycin A1 (Baf A1) (0.2 μM) and NH4Cl (20 mM) for 6 h, respectively. The cell lysates were then analyzed by immunoblot. i Protein lysates of WT, BECN1 KO and ATG5 KO 293 T cells transfected with plasmids encoding EV or HA-NEDD4, followed by SeV (MOI = 0.1) infection for 12 h were immunoblotted with indicated antibodies. For (a–i), similar results are obtained for three independent biological experiments.
To further determine whether NEDD4 degrades TBK1 via a ubiquitin-proteasome pathway or autolysosome pathway, we used pharmacological inhibitors and found the degradation of TBK1 induced by NEDD4 was blocked by 3-methyladenine (3MA), chloroquine (CQ), bafilomycin A1 (Baf A1), and NH4Cl, but not the proteasome inhibitor MG132 (Fig. 4h). In addition, the degradation of TBK1 triggered by NEDD4 was almost abrogated in BECN1 and ATG5 KO cells upon SeV infection (Fig. 4i). Consistently, cycloheximide (CHX)-chase assay results showed that the degradation of TBK1 were inhibited in BECN1 and ATG5 KO cells, comparing with WT 293 T cells (Supplementary Fig. 3c, d). Taken together, these results demonstrate that NEDD4 specifically degrades the active form of TBK1 during viral infection through autophagy.
NEDD4 enhances the recognition of TBK1 by cargo receptor NDP52
Accumulating evidence reveals that cargo receptors play critical roles in delivering substrates for selective autophagic degradation [26, 38]. Thus, we next attempted to identify the potential cargo receptor responsible for the autophagic degradation of TBK1. Coimmunoprecipitation assay suggested that TBK1 interacted with the cargo receptors p62, NDP52, and OPTN (Fig. 5a). Likewise, we found that NEDD4 also interacted with the cargo receptors p62, NDP52, and OPTN (Supplementary Fig. 4a). Furthermore, we observed that NEDD4 failed to inhibit the activation of type I IFN signaling in NDP52 KO cells, but not in SQSTM1 or OPTN KO cells (Fig. 5b and Supplementary Fig. 4b). In addition, we found that NEDD4 could promote the association between TBK1 and NDP52, but not p62 or OPTN (Supplementary Fig. 4c, d, e). We moved on to validate the role of NEDD4 in association between TBK1 and NDP52. Since NEDD4 significantly promotes the degradation of TBK1, the autophagy inhibitor Baf A1 was used to inhibit the reduction of TBK1 for better assessing the association of TBK1 with cargo receptors in coimmunoprecipitation analysis. Consistently, NEDD4 knockdown remarkably attenuated the association of endogenous TBK1 and NDP52 (Fig. 5c). Together, these data suggest that NEDD4 might mediate the NDP52-directed selective autophagic degradation of TBK1.
a 293 T cells were transfected with vectors encoding HA-TBK1, and indicated Flag-tagged cargo receptors, followed by immunoprecipitation with anti-Flag beads and immunoblot analysis with anti-HA. b Luciferase activity in 293 T wild-type (WT), SQSTM1 knockout (KO), NDP52 KO or OPTN KO cells, transfected with an ISRE (up) or IFN-β (down) promoter-driven luciferase reporter, together with plasmid encoding empty vector (EV) or expression vector of HA-NEDD4 after SeV (MOI = 0.1) infection for 12 h. NS: not significant. c A549 cells were transfected with scrambled (Scr) siRNA or NEDD4 siRNAs, followed by SeV (MOI = 0.1) infection for 12 h. Protein lysates were harvested after bafilomycin A1 (Baf A1) (0.2 μM) treatment (6 h) for immunoprecipitation using anti-TBK1 antibody and immunoblot using indicated antibodies. d Protein lysates of WT and NDP52 KO 293 T cells transfected with plasmids encoding EV or HA-NEDD4, followed by SeV (MOI = 0.1) infection for 12 h were immunoblotted with indicated antibodies. e Immunoblot analysis of protein extracts of WT and NDP52 KO 293 T cells treated with cycloheximide (CHX) (100 μg/mL) for indicated time points. f Coimmunoprecipitation and immunoblot analysis of protein lysates of 293 T cells transfected with vectors expressing WT Flag-NDP52 or Flag-NDP52 DR/CK mutant, followed by SeV (MOI = 0.1) infection for 12 h. Protein lysates were harvested after Baf A1 (0.2 μM) treatment (6 h) for immunoprecipitation and immunoblot using indicated antibodies. g Protein lysates of WT and NDP52 KO 293 T cells transfected with plasmids encoding EV or HA-NEDD4, together with WT NDP52 or its DR/CK mutant, were immunoblotted with indicated antibodies. In (b), all error bars, mean values ± SEM, P values were determined by unpaired two-tailed Student’s t test of n = 3 independent biological experiments, *** P < 0.001. For (a, c–g), similar results are obtained for three independent biological experiments.
We next investigated whether NDP52 is involved in autophagic degradation of TBK1. Indeed, we found NEDD4 failed to promote the degradation of TBK1 in NDP52 KO cells (Fig. 5d). Furthermore, CHX-chase assay result showed that the degradation rate of TBK1 in NDP52 KO cells was slower, comparing with WT cells (Fig. 5e). It has been well documented that NDP52 mainly recognizes ubiquitinated substrates for autophagic degradation [26, 39, 40]. Since NDP52 recognizes and delivers substrates to autophagosomes through its ubiquitin-associated (UBA) domain [41], we moved on to identify whether the UBA domain is essential for the recognition of TBK1. We generated NDP52 D439R/C443K (DR/CK) mutant, which has folding defects in the UBA domain [42], and observed that SeV infection increased the interaction of TBK1 and WT NDP52, whereas the NDP52 DR/CK mutant bound to TBK1 with a much lower affinity (Fig. 5f). Furthermore, we overexpressed NEDD4 with WT NDP52 or NDP52 DR/CK mutant in NDP52 KO cells and found that NEDD4 failed to promote the degradation of TBK1 after virus infection when restored with the NDP52 DR/CK mutant in NDP52 KO cells (Fig. 5g). These results indicate that NDP52 functions as cargo receptor to mediate the autophagic degradation of ubiquitinated TBK1.
NEDD4 increases the K27-linked poly-ubiquitination of TBK1
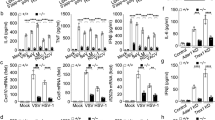
We then investigated whether NEDD4 could directly ubiquitinate TBK1, and found that NEDD4 depletion remarkably decreased the poly-ubiquitination of TBK1 (Fig. 6a). Furthermore, we found that NEDD4 specifically increased K27-linked (K27-only ubiquitin mutant) poly-ubiquitination of TBK1, but not the ubiquitination of TBK1 with other ubiquitin linkages in an overexpression system (Fig. 6b). To further confirm that NEDD4 physiologically mediates K27-linked poly-ubiquitination of TBK1, we performed MS analysis of ubiquitinated TBK1 from cells with SeV infection and found that the K27-linked poly-ubiquitinated TBK1 was readily detected after SeV infection, which was hardly detected without viral infection (Fig. 6c). Likewise, we found that TBK1 underwent the K27-linked poly-ubiquitination around 12 h post viral infection, which could be markedly enhanced by overexpression of NEDD4 (Fig. 6d). Consistently, depletion of NEDD4 resulted in decreased K27-linked poly-ubiquitination of TBK1 during SeV and VSV infection (Fig. 6e and Supplementary Fig. 5). Furthermore, we observed that NEDD4 mediated the ubiquitination and degradation of TBK1 in a catalytic activity-dependent manner (Fig. 6f, g). Taken together, these results suggest that NEDD4 is responsible for promoting the K27-linked poly-ubiquitination of TBK1.
a A549 cells were transfected with scrambled (Scr) siRNA or NEDD4 siRNAs, followed by SeV (MOI = 0.1) infection for 12 h. Protein lysates were harvested after bafilomycin A1 (Baf A1) (0.2 μM) treatment (6 h) for immunoprecipitation using anti-TBK1 antibody and immunoblot using indicated antibodies. b Lysates of 293 T cells transfected with plasmids expressing Flag-TBK1 and HA-tagged ubiquitin (Ub) or its indicated mutants, together with empty vector (EV) or expression vector of Myc-NEDD4 in the presence of Baf A1 (0.2 μM) for 6 h, were immunoprecipitated with anti-Flag beads and immunoblotted with anti-HA antibody. c Mass spectrometry analysis of a peptide derived from ubiquitinated Flag-TBK1. d Protein lysates of 293 T cells transfected with plasmids expressing HA-K27-Ub, together with EV or expression vector of Myc-NEDD4, followed by SeV (MOI = 0.1) infection for various time points (above lanes) were subjected to immunoprecipitation with anti-TBK1 antibody and immunoblot analysis with indicated antibodies. e A549 cells were transfected with Scr siRNA or NEDD4 siRNAs, followed by SeV (MOI = 0.1) infection for 12 h and Baf A1 (0.2 μM) treatment for 6 h. Protein lysates were immunoprecipitated using anti-TBK1 antibody and immunoblotted with anti-K27-Ub antibody. f 293 T cells were transfected with vectors for WT HA-NEDD4 or its HA/CA mutant, followed by SeV (MOI = 0.1) infection for 12 h. Protein lysates were immunoprecipitated with anti-TBK1 antibody and immunoblotted with indicated antibodies. g Coimmunoprecipitation and immunoblot analysis of 293 T cells transfected with Flag-TBK1 and HA-K27-Ub, together with vectors for WT HA-NEDD4 or its HA/CA mutant. Protein lysates were harvested after bafilomycin A1 (Baf A1) (0.2 μM) treatment (6 h) for immunoblotted with indicated antibodies. Samples in (a, b, and d–g) were incubated for 5 min with 1% SDS before immunoprecipitation, similar results are obtained for three independent biological experiments.
K344 is critical for the selective autophagic degradation of TBK1
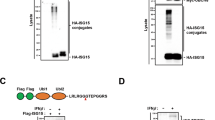
To gain insight into the mechanism of NEDD4 in regulating TBK1 ubiquitination, we generated three truncations of TBK1 and found that NEDD4 could promote the degradation of TBK1 mutant containing the kinase domain (KD) and ubiquitin-like domain (ULD), but not the mutants containing only KD domain or coiled-coil (CC) domain (Fig. 7a). We further found that NEDD4 could increase the K27-linked poly-ubiquitination of TBK1 mutant containing KD and ULD domains (Fig. 7b). These data indicate that the ULD domain of TBK1 is responsible for NEDD4-mediated degradation and ubiquitination of TBK1.
a Immunoblot analysis of protein extracts of 293 T cells transfected with wild-type (WT) or deletion mutants of Flag-TBK1 plasmids, together with empty vector (EV) or expression vector for HA-NEDD4. b 293 T cells were transfected with vectors expressing WT or deletion mutant of Flag-TBK1(KD + ULD), together with HA-K27-linked ubiquitin (Ub) and EV or expression vector for HA-NEDD4. Protein lysates were harvested after bafilomycin A1 (Baf A1) (0.2 μM) treatment (6 h) for immunoprecipitation and immunoblot using indicated antibodies. c Mass spectrometry analysis to show that K344 of TBK1 is conjugated with ubiquitin. d Coimmunoprecipitation and immunoblot analysis of protein extracts of 293 T cells transfected with vectors expressing WT Flag-TBK1 or its K344R mutant, together with HA-K27-Ub and EV or vector for Myc-NEDD4, in the presence of Baf A1 (0.2 μM) treatment for 6 h. e 293 T cells were transfected with plasmids expressing WT Flag-TBK1 or its indicated mutants, together with plasmid for HA-NDP52, in the presence of Baf A1 (0.2 μM) treatment for 6 h, followed by immunoprecipitated with anti-Flag beads and immunoblotted with anti-HA antibody. f Immunoblot analysis of protein lysates of 293 T cells transfected with HA-NEDD4, together with plasmid expressing WT Flag-TBK1 or its indicated mutants. g Protein lysates of WT and TBK1 knockout (KO) 293 T cells transfected with plasmids encoding EV or HA-NEDD4, together with WT TBK1 or its K344R mutant, and followed by SeV (MOI = 0.1) infection for 12 h, were immunoblotted with indicated antibodies. h RT–PCR analysis of indicated gene expression in TBK1 KO 293 T cells transfected with plasmids encoding EV or HA-NEDD4, together with WT TBK1 or its K344R mutant, and followed by SeV (MOI = 0.1) infection for 12 h. In (h), all error bars, mean values ± SEM, P values were determined by unpaired two-tailed Student’s t test of n = 3 independent biological experiments, ***P < 0.001. Samples in (b and d) were incubated for 5 min with 1% SDS before immunoprecipitation. For (a, b and d–g), similar results are obtained for three independent biological experiments.
We next investigated the possible NEDD4-mediated ubiquitination sites on ULD domain of TBK1. Ubiquitinated TBK1 from cells after SeV infection was precipitated and subjected to MS analysis. The MS data showed that the K344 residue on TBK1 was ubiquitinated (Fig. 7c). To further confirm that NEDD4 mediates the ubiquitination of TBK1 at K344, we identified four lysine sites within ULD domain of TBK1 and mutated them from K to R. Ubiquitination assay showed that only TBK1 K344R mutant displayed reduced K27-linked poly-ubiquitination (Supplementary Fig. 6a). Meanwhile, we found that NEDD4 could not promote the K27-linked poly-ubiquitination of TBK1 K344R mutant (Fig. 7d). Moreover, we found that TBK1 K344R mutant failed to interact with NDP52 (Fig. 7e), and NEDD4 failed to promote the degradation of TBK1 K344R mutant (Fig. 7f). We next determined whether K27-linked poly-ubiquitination on K344 functions as a degradation signal for TBK1 by a CHX-chase assay, and found that the degradation rate of TBK1 K344R mutant was slower, comparing with WT TBK1 (Supplementary Fig. 6b, c). These results suggest that K344 on TBK1 is critical for NDP52-mediated autophagic degradation of TBK1.
To address the functional importance of ubiquitination on K344 site of TBK1 in type I IFN signaling, we performed luciferase reporter assay and found that TBK1 K344R mutant retained the ability to induce luciferase activity in the presence of NEDD4 (Supplementary Fig. 6d). Furthermore, we overexpressed NEDD4 with WT TBK1 or TBK1 K344R mutant in TBK1 KO cells and found that NEDD4 failed to promote the degradation of TBK1 after virus infection when restored with the TBK1 K344R mutant in TBK1 KO cells, as well as phosphorylation of TBK1 and IRF3 (Fig. 7g). Consistently, the NEDD4-mediated decrease of transcription of IFNB, ISG54, and ISG56 by SeV infection, was abrogated in TBK1 K344R mutant-reconstituted cells (Fig. 7h and Supplementary Fig. 6e). Collectively, these results suggest that NEDD4 specifically promotes the K27-linked poly-ubiquitination of TBK1 at K344, which serves as a degradation signal for NDP52-mediated selective autophagy (Supplementary Fig. 6f).
Discussion
In the present study, we identified a novel role for NEDD4 in the negative regulation of antiviral immunity by showing that NEDD4 targets activated TBK1 for autophagic degradation through promoting its K27-linked poly-ubiquitination. Ectopic expression of NEDD4 inhibited the activation of type I IFN signaling and enhanced the infection of SeV and VSV. These results are consistent with previous reports that NEDD4 promotes virus infection [32]. Interestingly, NEDD4L, the close relative member of NEDD4 in NEDD4 family, was recently reported to promote antiviral immunity through mediating the K29-linked cysteine ubiquitination of TRAF3 [43]. These findings suggest different members of NEDD4 family might play distinct roles in innate antiviral immunity by controlling diverse ubiquitination types of their substrates. Further investigations are needed to clarify how different NEDD4 family members cooperate to orchestrate antiviral immunity.
Ubiquitination has been reported to be a critical post-translational modification to affect TBK1 function. K63-linked poly-ubiquitination promotes TBK1 activation in response to virus or lipopolysaccharide, thus inducing type I IFN signaling [44, 45], while K33-linked poly-ubiquitination competes with K48-linked poly-ubiquitination on TBK1 to maintain the half-life of TBK1 protein [14]. Conversely, K48-linked poly-ubiquitination of TBK1 occurs within 8 h post viral infection and results in its proteasomal degradation, to inhibit type I IFN signaling at the early stage of viral infection [10, 13, 14]. In the present study, we identified K27-linked poly-ubiquitination promotes autophagic degradation of activated TBK1 at the later stage of viral infection (around 12 h post infection), to avoid excessive type I IFN activation. These results indicate that different types of ubiquitination orchestrate to control TBK1 activity and stability through distinct degradation pathways during the whole process of viral infection.
TBK1 undergoes phosphorylation after virus infection, which is a required step for its activation as well as type I IFNs production. However, persistent TBK1 phosphorylation might cause autoimmune disorder, thus its activity and stability must be strictly controlled. Here, we found NEDD4 specifically promoted the degradation of phosphorylated TBK1. Importantly, NEDD4-induced degradation of TBK1 was inhibited by autophagy inhibitors, but not the proteasome inhibitor MG132. Moreover, NEDD4-mediated K27-linked poly-ubiquitination of TBK1 at K344 modulated the degradation of TBK1 through autolysosome pathway. Thus, NEDD4 functions as a molecular brake to prevent uncontrolled TBK1 activation through autophagy during viral infection.
Autophagy plays a critical role in the regulation of innate antiviral immune responses and host immunity. Nonetheless, the mechanisms underlying the crosstalk between selective autophagy and type I IFN signaling remain poorly understood. Selective autophagy requires the labeling of cargoes with degradation signals which can be recognized by cargo receptors, and then delivers the substrates to the autophagic membrane through their LC3-interacting region (LIR) [46]. Our recent studies demonstrated that the K27-linked poly-ubiquitin chains attached to the antiviral adaptor MAVS and IRF3 also serve as a degradation signal recognized by NDP52 in human cells [25, 38]. However, whether NDP52 recognizes the kinase of type I IFN signaling via K27-linked poly-ubiquitin was unclear. Here, we found that the K27-linked poly-ubiquitin chains on activated TBK1 could also be recognized by cargo receptor NDP52, leading to the autophagic degradation of ubiquitinated TBK1. Notably, K27-linked poly-ubiquitin chains might be a specific signal for NDP52-mediated selective autophagy in regulating innate immunity. Further studies are needed to investigate the specific ubiquitin signals for different cargo receptors.
In summary, we identified NEDD4 as an important regulator of antiviral immunity by promoting the autophagic degradation of TBK1. NEDD4 catalyzes the K27-linked poly-ubiquitination on TBK1 at K344, and facilitates the recognition and selective autophagic degradation of TBK1 via cargo receptor NDP52. Overall, our findings have identified a previously unrecognized role for NEDD4 in the homeostasis of innate immune signaling and have provided molecular insight into the mechanisms by which NDP52-mediated selective autophagy targets activated TBK1 for degradation.
Materials and methods
Cell lines and culture conditions
HEK293T and A549 cells obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) were maintained in DMEM medium (Gibco) with 10% (vol/vol) fetal bovine serum (Gibco) and 1% L-glutamine (Gibco). Peripheral blood samples from healthy individuals were collected at the Sun Yat-sen University Cancer Center according to the guidelines of the Ethics Committee of Sun Yat-sen University (GZR2013-040). Human peripheral blood mononuclear cells (PBMCs) were isolated from the blood of healthy donors. The use of PBMCs was in compliance with institutional ethics guidelines and approved protocols of Sun Yat-sen University. PBMCs were maintained in RPMI-1640 medium (Gibco) 10% fetal bovine serum. All cells were incubated at 37 °C incubator with 5% CO2.
Plasmids and transfection
Plasmids for NEDD4 and its mutants were cloned into the pcDNA3.1 vector for transient expression. HEK293T transfection was performed using Lipofectamine 2000 (Invitrogen) according to procedures recommended by the manufacturer. Chemically synthesized 21-nucleotide siRNA duplexes were obtained from TranSheepBio and transfected using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions. The sequences of target siRNAs are as follows:
siNEDD4-1: 5ʹ-GGCGAUUUGUAAACCGAAUTT-3ʹ;
siNEDD4-2: 5ʹ-GCAAGACAAGAGAUGAUUUTT-3ʹ;
siNEDD4-3: 5ʹ-GAACGACUACUUGGACAAATT-3ʹ;
Scrambled siRNA: 5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ.
Protein degradation inhibition assays
MG132 (10 μM) was used to inhibit proteasome-mediated protein degradation. 3MA (10 mM), CQ (50 μM), bafilomycin A1 (Baf A1) (0.2 μM) or NH4Cl (20 mM) was used to inhibit autolysosome- or lysosome-mediated protein degradation.
Antibodies and reagents
Monoclonal anti-Flag M2-peroxidase (A8592), monoclonal anti-β-actin antibody produced in mouse AC-74 (A2228), and anti-Flag M2 affinity gel (A2220) were purchased from Sigma. Anti-Myc Mouse Antibody (Agarose Conjugated) (M20012) was purchased from Abmart. Anti-MYC-horseradish peroxidase (11814150001) and anti-HA-peroxidase (high affinity from rat immunoglobulin G1) (12013819001) were purchased from Roche. ATG5 antibody (2630 S), Beclin-1 antibody (3738 S), IKKεantibody (2690 S), TBK1/NAK antibody (3013 S), phospho-TBK1/NAK (Ser172, D52C2) rabbit mAb (5483 S), and phospho-IRF3 (Ser396, 4D4G) rabbit mAb (4947 S) were purchased from Cell Signaling Technology. Anti-ubiquitin (sc-8017), anti-NEDD4-1 (sc-25508), IRF3 antibody (FL-425) (sc-9082), and TBK1 antibody (L-15) (sc-9910) were purchased from Santa Cruz Biotechnology. Anti-K27-Ub-HRP (ab181537) was purchased from Abcam. CALCOCO2 polyclonal antibody (12229-1-AP) and p62/SQSTM1 polyclonal antibody (18420-1-AP) were purchased from Proteintech Group. CF568 donkey anti-goat IgG (H + L) (20106-1) was purchased from Biotium. Alexa Fluor 488 goat anti-Rabbit IgG (H + L) (A-11034) was purchased from Thermofisher. Protein marker (M221-01) was purchased from GenStar.
Generation of NEDD4-inducible and knockout cell lines
The lentiviral vectors (15 μg) were co-transfected with 5 μg of an expression plasmid for the vesicular stomatitis virus G protein into the HEK293T cells. The medium was changed the following day and the viral containing supernatant was collected 48 h after transfection, filtered through a 0.45 μm filter and subsequently used to infect cells with polybrene (8 μg/mL). A549 cells were infected by incubation with lentivirus-containing supernatant for 48 h. For NEDD4 ectopic expression, lentiviral particles were produced by transfecting HEK293T cells with FG-EH-DEST-NEDD4, VSGV and Δ8.9. A549 cells were infected by incubation with lentivirus-containing supernatant for 48 h. To generate NEDD4 KO, target sequences (NEDD4 guide: 5ʹ-GTATATGGTCTCTCCAATCT-3ʹ) were cloned into pLentiCRISPRv2 by cutting with BsmBI. Infected cells were treated with 1 μg/mL puromycin for 48 h to enrich transfected cells, which were then diluted and placed into 96-well plates for single colonies. The gRNA for TBK1, SQSTM1, NDP52, and OPTN were previously reported [47].
Immunoprecipitation and immunoblot analysis
For immunoprecipitation, whole-cell extracts were prepared after transfection or stimulation with appropriate ligands, followed by incubation overnight with the appropriate antibodies plus Protein A/G beads (Pierce) or anti-Flag agarose beads (Sigma). Beads were then washed five times with low-salt lysis buffer (50 mM HEPES, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 1.5 mM MgCl, and 1% Triton X-100), and immunoprecipitates were eluted with 2×SDS Loading Buffer and resolved by SDS-PAGE. For ubiquitination assays in cultured cells, the cells were lysed with low-salt lysis buffer and the supernatants were denatured at 95 °C for 5 min in the presence of 1% SDS. The denatured lysates were diluted with lysis buffer to reduce the concentration of SDS below 0.1% followed by immunoprecipitation (denature-IP) with the indicated antibodies. Proteins were transferred to PVDF membranes (Bio-Rad) and further incubated with the appropriate antibodies. Immobilon Western Chemiluminescent HRP Substrate (Millipore) was used for protein detection.
Fluorescence microscopy
Cells were cultured on Glass Bottom culture dishes (Nest Scientific) and directly observed as previously described [24]. For examination by immunofluorescence microscopy, cells were fixed with 4% paraformaldehyde for 15 min, and then permeabilized in methyl alcohol for 10 min at −20 °C. After washing with PBS for 3 times, cells were blocked in 5% fetal goat serum (Boster Biological) for 1 h, and then incubated with primary antibodies diluted in 10% bull serum albumin (Sigma) overnight. The cells were washed, and followed by a fluorescently labeled secondary antibody (Alexa Fluor 488 goat anti-Rabbit IgG (H + L) and CF568 donkey anti-goat-IgG). Confocal images were examined using a Leica DMI3000 B microscopy (DMI3000 B, Leica) or Leica TCS-SP5 confocal microscope (TCS-SP5, Leica) equipped with a × 100 NA oil-immersion objective.
Virus infection
SeV and VSV-eGFP were kindly provided by Dr. Xiaofeng Qin (Suzhou Institute of Systems Medicine). SeV and VSV-eGFP were titered on Vero cells. Virus titers were measured by means of 50% of the tissue culture’s infectious dose (TCID50).
Quantitative RT-PCR
Total RNA was extracted from cells using the Trizol reagent (Invitrogen) according to the manufacturer’s instructions. For RT-PCR analysis, cDNA was generated with HiScript® II Q RT SuperMix for qPCR (+gDNA wiper) (Vazyme, R223-01) and was analyzed by PCR using the 2×Taq PCR StarMix (GenStar). The sequences of primers are as follows:
TBK1: Forward: 5ʹ- TCATCTTAGGAAACAGTTAT-3ʹ,
Reverse: 5ʹ- GTAAACATTTTCTGAGGC -3ʹ;
RPL13A: Forward: 5ʹ-GCCATCGTGGCTAAACAGGTA-3ʹ,
Reverse: 5ʹ-GTTGGTGTTCATCCGCTTGC-3ʹ;
IFNB: Forward: 5ʹ-GATGAACTTTGACATCCCTGAG-3ʹ,
Reverse: 5ʹ-TCAACAATAGTCTCATTCCAGC-3ʹ;
ISG54: Forward: 5ʹ-TATTGGTGGCAGAAGAGGAAGA-3ʹ,
Reverse: 5ʹ-CAGGTGAAATGGCATTTTAGTT-3ʹ;
ISG56: Forward: 5ʹ-TCAGGTCAAGGATAGTCTGGAG-3ʹ,
Reverse: 5ʹ-AGGTTGTGTATTCCCACACTGTA-3ʹ;
SeV P: Forward: 5ʹ-TGTTATCGGATTCCTCGACGCAGTC-3ʹ,
Reverse: 5ʹ-TACTCTCCTCACCTGATCGATTATC-3ʹ;
VSV G: Forward: 5ʹ- CAAGTCAAAATGCCCAAGAGTCACA-3ʹ,
Reverse: 5′- TTTCCTTGCATTGTTCTACAGATGG-3′.
Luciferase and reporter assays
Cells were plated in 24-well plates and transfected with plasmids encoding the ISRE/IFNB luciferase reporter (firefly luciferase; 30 ng) and pRL-TK (Renilla luciferase; 10 ng), together with different plasmids (100 ng). The same number of cells (1 × 105) treated with SeV, VSV or IC poly(I:C) stimulation for indicated times were collected and luciferase activity was measured with Dual-Luciferase Assay (Promega, E1910) with a Luminoskan Ascent luminometer (Thermo Fisher Scientific). Reporter gene activity was determined by normalization of the firefly luciferase activity to Renilla luciferase activity. The values were means ± SD of 3 independent transfections performed in parallel.
Mass spectrometry analysis
Flag-tagged TBK1 was stably overexpressed in 293 T cells. The cell lysates were immunoprecipitated with anti-Flag beads. The beads were washed three times with 1x TBS and eluted with 1x loading buffer. Eluted proteins were identified with a gel-based liquid chromatography-tandem mass spectrometry (Gel-LS-MS/MS) approach. Mass spectra were analyzed using MaxQuant software (version 1.5.3.30) with the UniProtKB/Swiss-Prot human database. To detect ubiquitinated TBK1, Flag-tagged TBK1 immunoprecipitates were prepared from whole-cell lysates and resolved on 6% SDS-PAGE gels. The protein band where Flag-TBK1 accumulated were excised and subjected to Gel-LS-MS/MS sequencing and data analysis. The ubiquitinated peptides were analyzed using MASCOT engine (Matrix Science, London, UK; version 2.2) embedded into Proteome Discoverer 1.4 (Thermo Electron, San Jose, CA.).
RNA-seq analysis
Total RNA was extracted by Trizol (Invitrogen, 10296010), and the RNA sequencing was performed using the Illumina platform by Sangon Biotech Company. Quality control of the fastq data was performed with FASTQC. High quality reads were aligned to the human reference genome (mm10) using histat2 [48]. The aligned SAM file was converted into sorted BAM files by SAMtools, and the HTSeq-count program was used to count the total number of reads that are mapped to the genome. The differential gene expression analysis was performed by DESeq2 R package. Gene enrichment was analyzed by clusterProfiler R package.
Statistical analysis
Data are represented as mean ± SEM unless otherwise indicated, and Student’s t test was used for all statistical analyses with the GraphPad Prism 8 software. Differences between two groups were considered significant when P value was less than 0.05.
Data availability
Data supporting the present study are available from the corresponding author upon reasonable request.
References
Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–20.
Choi Y, Bowman JW, Jung JU. Autophagy during viral infection—a double-edged sword. Nat Rev Microbiol. 2018;16:341–54.
Tian Y, Wang M-L, Zhao J. Crosstalk between autophagy and type I interferon responses in innate antiviral immunity. Viruses. 2019;11:132.
Chen K, Liu J, Cao X. Regulation of type I interferon signaling in immunity and inflammation: a comprehensive review. J Autoimmun. 2017;83:1–11.
Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal. 2012;5:ra20.
Liu J, Qian C, Cao X. Post-translational modification control of innate immunity. Immunity. 2016;45:15–30.
Saul VV, Niedenthal R, Pich A, Weber F, Schmitz ML. SUMO modification of TBK1 at the adaptor-binding C-terminal coiled-coil domain contributes to its antiviral activity. Biochim Biophys Acta. 2015;1853:136–43.
Li X, Zhang Q, Ding Y, Liu Y, Zhao D, Zhao K, et al. Methyltransferase Dnmt3a upregulates HDAC9 to deacetylate the kinase TBK1 for activation of antiviral innate immunity. Nat Immunol. 2016;17:806–15.
Lei C-Q, Zhong B, Zhang Y, Zhang J, Wang S, Shu H-B. Glycogen synthase kinase 3 beta regulates IRF3 transcription factor-mediated antiviral response via activation of the kinase TBK1. Immunity. 2010;33:878–89.
Cui J, Li Y, Zhu L, Liu D, Songyang Z, Wang HY, et al. NLRP4 negatively regulates type I interferon signaling by targeting the kinase TBK1 for degradation via the ubiquitin ligase DTX4. Nat Immunol. 2012;13:387–95.
Zhang M, Wang L, Zhao X, Zhao K, Meng H, Zhao W, et al. TRAF-interacting protein (TRIP) negatively regulates IFN-beta production and antiviral response by promoting proteasomal degradation of TANK-binding kinase 1. J Exp Med. 2012;209:1703–11.
Zheng Q, Hou J, Zhou Y, Yang Y, Xie B, Cao X. Siglec1 suppresses antiviral innate immune response by inducing TBK1 degradation via the ubiquitin ligase TRIM27. Cell Res. 2015;25:1121–36.
Deng M, Tam JW, Wang L, Liang K, Li S, Zhang L, et al. TRAF3IP3 negatively regulates cytosolic RNA induced anti-viral signaling by promoting TBK1 K48 ubiquitination. Nat Commun. 2020;11:2193.
Lin M, Zhao Z, Yang Z, Meng Q, Tan P, Xie W, et al. USP38 inhibits type I interferon signaling by editing TBK1 ubiquitination through NLRP4 signalosome. Mol Cell. 2016;64:267–81.
Abe T, Lee A, Sitharam R, Kesner J, Rabadan R, Shapira SD. Germ-cell-specific inflammasome component NLRP14 negatively regulates cytosolic nucleic acid sensing to promote fertilization. Immunity. 2017;46:621–34.
Ma Y, Galluzzi L, Zitvogel L, Kroemer G. Autophagy and cellular immune responses. Immunity. 2013;39:211–27.
Clarke AJ, Simon AK. Autophagy in the renewal, differentiation and homeostasis of immune cells. Nat Rev Immunol. 2019;19:170–83.
Jang YJ, Kim JH, Byun S. Modulation of autophagy for controlling immunity. Cells. 2019;8:138.
Deretic V, Levine B. Autophagy balances inflammation in innate immunity. Autophagy. 2018;14:243–51.
Kumar S, Gu Y, Abudu YP, Bruun JA, Jain A, Farzam F, et al. Phosphorylation of syntaxin 17 by TBK1 controls autophagy initiation. Dev Cell. 2019;49:130–44. e136
Richter B, Sliter DA, Herhaus L, Stolz A, Wang C, Beli P, et al. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc Natl Acad Sci USA. 2016;113:4039–44.
Du Y, Duan T, Feng Y, Liu Q, Lin M, Cui J, et al. LRRC25 inhibits type I IFN signaling by targeting ISG15-associated RIG-I for autophagic degradation. EMBO J. 2018;37:351–66.
Xian H, Yang S, Jin S, Zhang Y, Cui J. LRRC59 modulates type I interferon signaling by restraining the SQSTM1/p62-mediated autophagic degradation of pattern recognition receptor DDX58/RIG-I. Autophagy. 2020;16:408–18.
Chen M, Meng Q, Qin Y, Liang P, Tan P, He L, et al. TRIM14 inhibits cGAS degradation mediated by selective autophagy receptor p62 to promote innate immune responses. Mol Cell. 2016;64:105–19.
Jin S, Tian S, Luo M, Xie W, Liu T, Duan T, et al. Tetherin suppresses type I interferon signaling by targeting MAVS for NDP52-mediated selective autophagic degradation in human cells. Mol Cell. 2017;68:308–22.
He X, Zhu Y, Zhang Y, Geng Y, Gong J, Geng J, et al. RNF34 functions in immunity and selective mitophagy by targeting MAVS for autophagic degradation. EMBO J. 2019;38:e100978.
Prabakaran T, Bodda C, Krapp C, Zhang B-C, Christensen MH, Sun C, et al. Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. EMBO J. 2018;37:e97858.
Yang Q, Liu T-T, Lin H, Zhang M, Wei J, Luo W-W, et al. TRIM32-TAX1BP1-dependent selective autophagic degradation of TRIF negatively regulates TLR3/4-mediated innate immune responses. PLoS Pathog. 2017;13:e1006600.
Kimura T, Jain A, Choi SW, Mandell MA, Schroder K, Johansen T, et al. TRIM-mediated precision autophagy targets cytoplasmic regulators of innate immunity. J Cell Biol. 2015;210:973–89.
Li Y, Zhang L, Zhou J, Luo S, Huang R, Zhao C, et al. Nedd4 E3 ubiquitin ligase promotes cell proliferation and autophagy. Cell Prolif. 2015;48:338–47.
Huang X, Chen J, Cao W, Yang L, Chen Q, He J, et al. The many substrates and functions of NEDD4-1. Cell Death Dis. 2019;10:904.
Chesarino NM, McMichael TM, Yount JS. E3 ubiquitin ligase NEDD4 promotes influenza virus infection by decreasing levels of the antiviral protein IFITM3. PLoS Pathog. 2015;11:e1005095.
Xie W, Jin S, Wu Y, Xian H, Tian S, Liu D-A, et al. Auto-ubiquitination of NEDD4-1 recruits USP13 to facilitate autophagy through deubiquitinating VPS34. Cell Rep. 2020;30:2807–19.
Pei G, Buijze H, Liu H, Moura-Alves P, Goosmann C, Brinkmann V, et al. The E3 ubiquitin ligase NEDD4 enhances killing of membrane-perturbing intracellular bacteria by promoting autophagy. Autophagy. 2017;13:2041–55.
Lin Q, Dai Q, Meng H, Sun A, Wei J, Peng K, et al. The HECT E3 ubiquitin ligase NEDD4 interacts with and ubiquitylates SQSTM1 for inclusion body autophagy. J Cell Sci. 2017;130:3839–50.
Malakhova OA, Zhang D-E. ISG15 inhibits Nedd4 ubiquitin E3 activity and enhances the innate antiviral response. J Biol Chem. 2008;283:8783–7.
Okumura A, Pitha PM, Harty RN. ISG15 inhibit Ebola VP40VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proc Natl Acad Sci USA. 2008;105:3974–9.
Wu Y, Jin S, Liu Q, Zhang Y, Ma L, Zhao Z, et al. Selective autophagy controls the stability of transcription factor IRF3 to balance type I interferon production and immune suppression. Autophagy. 2021;17:1379–92.
Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13:722–37.
Jo C, Gundemir S, Pritchard S, Jin YN, Rahman I, Johnson GVW. Nrf2 reduces levels of phosphorylated tau protein by inducing autophagy adaptor protein NDP52. Nat Commun. 2014;5:3496.
Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–96.
Xie X, Li F, Wang Y, Wang Y, Lin Z, Cheng X, et al. Molecular basis of ubiquitin recognition by the autophagy receptor CALCOCO2. Autophagy. 2015;11:1775–89.
Gao P, Ma X, Yuan M, Yi Y, Liu G, Wen M, et al. E3 ligase Nedd4l promotes antiviral innate immunity by catalyzing K29-linked cysteine ubiquitination of TRAF3. Nat Commun. 2021;12:1194.
Song G, Liu B, Li Z, Wu H, Wang P, Zhao K, et al. E3 ubiquitin ligase RNF128 promotes innate antiviral immunity through K63-linked ubiquitination of TBK1. Nat Immunol. 2016;17:1342–51.
Wang C, Chen T, Zhang J, Yang M, Li N, Xu X, et al. The E3 ubiquitin ligase Nrdp1 ‘preferentially’ promotes TLR-mediated production of type I interferon. Nat Immunol. 2009;10:744–52.
Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018;19:349–64.
Vargas JNS, Wang C, Bunker E, Hao L, Maric D, Schiavo G, et al. Spatiotemporal control of ULK1 activation by NDP52 and TBK1 during selective autophagy. Mol Cell. 2019;74:347–62.
Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11:1650–67.
Acknowledgements
We are grateful to Jiang Li and Daijia Huang (Sun Yat-sen University Cancer Center) for the kind gift of PBMCs.
Funding
This work was supported by the National Natural Science Foundation of China (31870862, 92042303, and 31970700), Science and Technology Planning Project of Guangzhou, China (201907010038), Guangdong Basic and Applied Basic Research Foundation (2020B1515120090), Guangdong Science and Technology Department (2020B1212060031), Natural Science Foundation of Guangdong Province, China (2021A1515012179), and China Postdoctoral Science Foundation (2020TQ0388 and 2020M683036).
Author information
Authors and Affiliations
Contributions
WX performed the experiments and analyzed the results. SJ, CZ, SY, YW, YZ and ZS provided technical help. JC initiated and designed the project and directed the research. WX and JC wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics statement
The use of PBMCs was in compliance with institutional ethics guidelines and approved protocols of Sun Yat-sen University.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by M. Piacentini
Rights and permissions
About this article
Cite this article
Xie, W., Jin, S., Zhang, C. et al. Selective autophagy controls the stability of TBK1 via NEDD4 to balance host defense. Cell Death Differ 29, 40–53 (2022). https://doi.org/10.1038/s41418-021-00833-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-021-00833-9
This article is cited by
-
The role of NEDD4 related HECT-type E3 ubiquitin ligases in defective autophagy in cancer cells: molecular mechanisms and therapeutic perspectives
Molecular Medicine (2023)
-
The role of TBK1 in cancer pathogenesis and anticancer immunity
Journal of Experimental & Clinical Cancer Research (2022)