Abstract
CD4/CD8 T-cell lineage differentiation is a key process in immune system development; however, a defined regulator(s) that converts the signal from T-cell receptor and co-receptor complexes into lineage differentiation remains unclear. Here, we show that Twist2 is a critical factor in CD4/CD8 thymocyte differentiation. Twist2 expression is differentially regulated by T-cell receptor signaling, leading to differentiation into the CD4 or CD8 lineage. Forced Twist2 expression perturbed CD4+ thymocyte differentiation while enhancing CD8+ thymocyte differentiation. Furthermore, Twist2 expression produced mature CD8+ thymocytes in B2m−/− mice, while its deficiency significantly impaired CD8+ cells in MHC class-II−/− and TCR transgenic mice, favoring CD8 T-cell differentiation. During CD8 lineage differentiation, Twist2 interacted with Runx3 to bind to the silencer region of the ThPOK locus, thereby blocking ThPOK expression. These findings indicate that Twist2 is a part of the transcription factor network controlling CD8 lineage differentiation.
Similar content being viewed by others
Introduction
Bone marrow-derived T-cell progenitors develop in the thymus through stages characterized by the expression of CD4 and CD8 co-receptors [1, 2]; CD4−CD8− (double-negative, DN), CD4+CD8+ (double-positive, DP), and CD4 single-positive (CD4 SP) or CD8 SP cells. DP thymocytes experience positive and negative selection depending on the interaction of their TCRs with the major histocompatibility complex (MHC) expressed on thymic stromal cells. Only a small fraction of DP thymocytes can differentiate into CD4 SP or CD8 SP cells.
The commitment of DP thymocytes between CD4 SP and CD8 SP lineages has been widely investigated [3,4,5,6]. It is generally accepted that matching MHC classes and TCR with a CD4 or CD8 co-receptor is critical for differentiation [7]. In addition, it appears that signal transduction from the TCR is crucial for CD4/CD8 lineage differentiation. Numerous genetic and biochemical studies have demonstrated that strong and persistent TCR signals lead to CD4 SP cells, while weak and transient TCR signals lead to CD8 SP cells [8, 9].
Several studies have revealed a few transcription factors that may link proximal TCR signaling to the CD4/CD8 lineage commitment process [10,11,12,13,14,15,16,17]. GATA3 and ThPOK reportedly play critical roles in CD4 SP differentiation. GATA3 deficiency severely perturbs CD4 SP development, whereas GATA3 overexpression inhibits maturation of CD8 SP cells [18, 19]. Similarly, ThPOK mutation results in complete disruption of CD4 SP development, whereas ThPOK overexpression in the thymus redirects MHC class-I-restricted thymocytes to the CD4 SP thymocyte lineage [10, 14]. ThPOK expression is controlled by several cis-acting elements [20, 21]. Among them, the ThPOK silencer is the most critical for helper lineage-specific expression of the ThPOK gene. The ThPOK silencer is occupied by the Runx complex and is regulated by Mazr and Tle, which bind to the region [15, 16, 20]. In addition, CD8 lineage commitment involves epigenetic sealing of the silencer of the ThPOK locus during a temporal developmental window [22]. Conversely, Runx3 plays important roles in CD8 SP differentiation. Runx3-deficient mice showed impaired CD8 SP development, whereas forced expression of Runx3 in thymocytes increased the proportion of CD8 SP mature thymocytes [12, 23]. Runx3 can suppress both CD4 co-receptor and ThPOK expression by directly binding to the silencer region of both genes [20, 24]. However, the Runx complex is bound to both the silencer and enhancer region of ThPOK throughout T-cell differentiation. Thus, differential regulation of ThPOK expression in CD4/CD8 lineage differentiation requires additional factor(s) besides Runx.
Twist2 is a basic helix-loop-helix transcription factor first identified as an E12 binding protein [25]. Twist2-deficient mice show perinatal death due to cachexia, as well as increased apoptotic bodies in the thymus and spleen [26]. It was also reported that Twist2 expression is regulated by TCR signaling; to balance the survival and death of thymocytes, Twist2 controls TCR-mediated apoptosis by regulating the expression of Nur77 and Nor-1 [27]. These results suggest that Twist2 plays an important role during lymphocyte development.
Here, we show that Twist2 is induced by TCR signaling and its expression is upregulated more under CD8 differentiation conditions than under CD4 differentiation conditions. Twist2 binds to the silencer region of the ThPOK gene by forming a complex with Runx3, and represses ThPOK expression and, thereby, represses CD4 T-cell differentiation and induces CD8 T-cell differentiation.
Results
Twist2 is upregulated by TCR stimuli
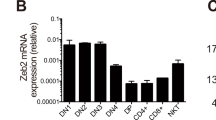
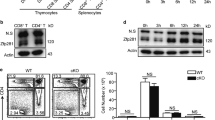
To gain insight into the role of Twist2 in thymocyte differentiation, thymocytes at various developmental stages were sorted by flow cytometry and Twist2 expression was analyzed by quantitative reverse transcription polymerase chain reaction (RT-qPCR) and western blotting. After the DP stage, Twist2 expression was increased at the CD4+CD8lo (CD4+CD8loCD69hiTCRβ+; simplified “CD4+CD8lo” hereafter) stage, and its expression was higher in CD8 SP cells than in CD4 SP cells at both the transcriptional and translational levels (Fig. 1a, b and Supplementary Fig. S1). To investigate whether such differential expression of Twist2 is observed during CD4- or CD8-SP-favored differentiation, Twist2 expression levels were measured in thymocytes collected from various genetically modified mice. RT-qPCR analysis indicated that Twist2 expression was higher in total thymocytes from mouse model favoring CD8 SP differentiation (see MHC II−/−) than in those from mouse model favoring CD4 SP differentiation (see B2m−/−) (Fig. 1c).
a Twist2 expression was analyzed by Q-PCR in the indicated thymocyte subsets from C57BL/6 mice (n = 3). b Nuclear extracts were prepared from DP, CD4 SP, and CD8 SP cells of C57BL/6 mice. Twist2 expression was determined by western hybridization using anti-Twist2 antibodies. Data are from one representative of three independent experiments. c Twist2 and Runx3 expression was analyzed by Q-PCR in total thymocytes from the indicated mice (n = 3). d Thymocytes from Tcra−/−mice were activated with 0.2 ng/mL (CD4 conditions) or 0.1 ng/mL PMA (CD8 conditions) and 0.2 μg/mL ionomycin for the indicated times. Twist2 expression was determined by Q-PCR (n = 3). e Numbers of total thymocytes from NLC and Twist2 transgenic mice. Each symbol indicates an individual mouse (error bars, ±S.E.M.). f Thymocyte differentiation profiles based on the indicated surface markers from NLC and Twist2 transgenic mice. g The ratio of TCRβ+CD24lo CD4 SP cells to CD8 SP cells from NLC and Twist2 transgenic mice. Each symbol indicates an individual mouse (error bars, ±S.E.M.). h Cell numbers of mature CD4 SP and CD8 SP thymocytes in NLC and Twist2 transgenic mice. i Numbers of total thymocytes from CD4-Cre Twist2+/+, CD4-Cre Twist2f/+, and CD4-Cre Twist2f/f mice. Each symbol indicates an individual mouse (error bars, ±S.E.M.). j Thymocyte differentiation profiles based on the indicated surface markers from the indicated mice. k The ratio of TCRβ+CD24lo CD4 SP cells to CD8 SP cells from the indicated mice. Each symbol indicates an individual mouse (error bars, ±S.E.M.). l Cell numbers of CD4 SP and CD8 SP from CD4-Cre Twist2+/+ and CD4-Cre Twist2f/f mice. *p < 0.05, **p < 0.005, ***p < 0.001 (two-tailed Student’s t test).
Since Twist2 expression was increased in CD4+CD8lo thymocytes and differentially regulated in the CD4 or CD8 SP lineages, we analyzed its expression during thymocyte selection processes. Thymocytes from Tcra−/− mice were activated to differentiate into either CD4 or CD8 lineages as described previously (0.1 PMA induces CD8 SP, and 0.2 PMA induces CD4 SP) [11], and Twist2 expression was determined by RT-qPCR. Twist2 expression was induced in cultured DP thymocytes activated under conditions that gave rise to either CD4 or CD8 SP cells. The induced expression of Twist2 showed a similar pattern in the early activation phase under both conditions, but showed distinct patterns at the final evaluation time point, similar to its expression in CD4 and CD8 SP cells (Fig. 1d). Overall, these results suggest that Twist2 expression is more upregulated under CD8 differentiation conditions.
CD4/CD8 SP thymocyte ratios are altered in Twist2 transgenic and Twist2 conditional knockout (cKO) mice
On the basis of the differential expression of Twist2 between CD4 and CD8 lineage cells, we investigated the possible roles of Twist2 during thymocyte CD4/CD8 lineage differentiation. To investigate the function of Twist2, Twist2 transgenic mice and Twist2 conditional knockout (cKO) mice were analyzed [27]. Interestingly, the thymi of Twist2 transgenic mice were visibly reduced in size, and the total number of thymocytes was consistently reduced (Fig. 1e). The ratio of TCRβ+CD24lo CD4 to CD8 SP was significantly decreased from 2.18 ± 0.08 (mean ± standard error of the mean [SEM]) in non-transgenic littermate control mice to 1.48 ± 0.12 in Twist2 transgenic mice (Fig. 1f, g). The number of CD4 SP cells was significantly reduced. Even though the percentage of CD8 SP cells was increased, its absolute cell number was decreased, which is due mainly to the severe reduction in the total number of thymocytes (Fig. 1h). It was reported that T-cell lineage-specific deletion of Twist2 slightly decreases total thymic cellularity (Fig. 1i). Compared with CD4-Cre Twist2+/+ mice, CD4-Cre Twist2f/f mice showed altered CD4 and CD8 SP differentiation profiles, with a decreased cell number and percentage of TCRβ+CD24lo thymocytes (Supplementary Fig. S2). In addition, the ratio of TCRβ+CD24lo CD4 SP to CD8 SP was significantly increased from 2.06 ± 0.132 in CD4-Cre Twist2+/+ mice to 3.14 ± 0.174 in CD4-Cre Twist2f/f mice, which is thought to result from a decrease in the number of CD8 SP cells (3.51 × 106 vs. 1.88 × 106 in control and Twist2 cKO, respectively) (Fig. 1j–l). These results suggest that Twist2 can regulate the CD4/CD8 thymocyte ratio.
Altered CD4/CD8 SP thymocyte differentiation in Twist2 transgenic and Twist2 cKO mice expressing transgenic TCR
Based on the differential expression of Twist2 during CD4 and CD8 SP thymocyte differentiation in vitro, as well as the altered CD4/CD8 SP thymocyte ratios in Twist2 transgenic and cKO mice, we further investigated the effect of Twist2 on thymocyte differentiation using TCR transgenic mice. Since DO-TCR is restricted to MHC class-II molecules, these transgenic mice preferentially generate CD4 SP cells [28]. In Rag−/− DO-TCR and Twist2 double transgenic mice, the total number of thymocytes was consistently reduced and the percentage (91.67% vs. 32.97% in control and double transgenic mice, respectively) and cell number (2.99 × 106 vs. 0.52 × 106 cells) of the CD4 SP population were significantly decreased compared with that of the control, whereas the corresponding percentage (2.02% vs. 17.67%) and cell number (0.07 × 106 vs. 0.27 × 106 cells) of the CD8 SP population were increased, resulting in a decreased relative DO+CD24lo CD4 to CD8 SP thymocyte ratio (Fig. 2a–c). In the periphery, the percentage and cell number of the CD4 T cells were also decreased, whereas the percentage and cell number of CD8 T cells were increased in Rag−/− DO-TCR Twist2 transgenic mice (Fig. 2b and Supplementary Fig. S3).
a Numbers of total thymocytes from Rag−/− DO-TCR transgenic mice and Rag−/− DO-TCR Twist2 transgenic mice. b CD4 and CD8α expression in total thymocytes, DO+CD24lo thymocytes, and DO11.10hi splenocytes from Rag−/− DO-TCR transgenic mice and Rag−/− DO-TCR Twist2 transgenic mice. c Frequency and cell number of mature CD4 SP and CD8 SP thymocytes in Rag−/− DO-TCR transgenic mice and Rag−/− DO-TCR Twist2 transgenic mice (error bars, ±S.E.M., n = 3). d Numbers of total thymocytes from Rag−/− HY-TCR CD4-Cre Twist2+/+ and Rag−/− HY-TCR CD4-Cre Twist2f/f mice. e CD4 and CD8α expression in total thymocytes, HY+CD24lo thymocytes, and T3.70hi splenocytes from Rag−/− HY-TCR CD4-Cre Twist2+/+ and Rag−/− HY-TCR CD4-Cre Twist2f/f mice. f Frequency and cell number of mature CD4 SP and CD8 SP thymocytes in Rag−/− HY-TCR CD4-Cre Twist2+/+ and Rag−/− HY-TCR CD4-Cre Twist2f/f mice (error bars, ±S.E.M., n = 3). *p < 0.05, **p < 0.005, ***p < 0.001, ****p < 0.0001 (two-tailed Student’s t test).
To investigate the loss of Twist2 function during thymocyte differentiation, we generated Twist2 cKO mice in the Rag−/− HY-TCR transgenic background, in which transgenic TCR was restricted to MHC class-I molecules [29]. The percentage of CD4 SP cells was slightly increased but not significantly, and the number of CD4 SP cells was similar to that of control mice. Interestingly, however, the frequency of the CD8 SP population was decreased in the Rag−/− HY-TCR CD4-Cre Twist2f/f mice compared with that of the control mice (72.33% vs. 89.53%, respectively). Overall thymocyte cellularity was reduced but not significantly in the Rag−/− HY-TCR CD4-Cre Twist2f/f mice compared with that of the control mice (Fig. 2d). The cell number and percentage of T3.70+CD24lo thymocytes were reduced, and consequently there was a significant reduction in mature CD8 SP cell numbers in Rag−/− HY-TCR CD4-Cre Twist2f/f mice compared with that of the control mice (0.90 × 106 vs 0.47 × 106, respectively) (Supplementary Fig. S4a and Fig. 2e, f). Also, in the periphery, the percentage and cell number of the CD8 T cells were decreased in Rag−/− HY-TCR CD4-Cre Twist2f/f mice (Fig. 2e and Supplementary Fig. S4b). These results collectively suggest that Twist2 is required for CD8 SP cell differentiation.
Twist2 affects CD4/CD8 lineage differentiation to support CD8 SP differentiation
Twist2 was shown to produced MHC class-II-restricted CD8 SP cells in Rag−/− DO-TCR transgenic mice; thus, we next tested whether Twist2 can generate CD8 SP cells in the absence of MHC class I. Compared with B2m−/− mice, B2m−/− Twist2 transgenic mice generated significant numbers of CD8 SP cells in the thymus, and the cell number and the percentage of TCRβ+ CD4 SP thymocytes were significantly decreased (Fig. 3a–c). In the periphery, the B2m−/− Twist2 transgenic mice displayed significant numbers of CD8 T cells (Supplementary Fig. S5a). The functional nature of the CD8 SP cells was confirmed by measuring the expression of Perforin1 in the thymocytes (Supplementary Fig. S5b).
a Numbers of total thymocytes from B2m−/− and B2m−/− Twist2 transgenic mice. b Thymocyte differentiation profiles based on CD4 and CD8α expression of total thymocytes, TCRβ+ thymocytes, and TCRβ+ splenocytes from B2m−/− and B2m−/− Twist2 transgenic mice. c Frequency and cell number of mature CD4 SP and CD8 SP thymocytes in B2m−/− and B2m−/− Twist2 transgenic mice (error bars, ±S.E.M., n = 3). d Numbers of total thymocytes from MHC class-II−/− CD4-Cre Twist2+/+ and MHC class-II−/− CD4-Cre Twist2f/f mice. e Thymocyte differentiation profiles based on CD4 and CD8α expression of total thymocytes, TCRβ+CD24lo thymocytes, and TCRβ+ splenocytes from MHC class-II−/− CD4-Cre Twist2+/+ and MHC class-II−/− CD4-Cre Twist2f/f mice. f Frequency and cell numbers of CD8 SP from MHC class-II−/− CD4-Cre Twist2+/+ and MHC class-II−/− CD4-Cre Twist2f/f mice (error bars, ±S.E.M., n = 3). *p < 0.05, **p < 0.005, ***p < 0.001 (two-tailed Student’s t test).
We also analyzed the effect of Twist2 deficiency on CD4/CD8 lineage differentiation in an MHC class-II-deficient background. Overall thymic cellularity and the cell number and percentage of TCRβ+CD24lo thymocytes were decreased in the MHC class-II−/− CD4-Cre Twist2f/f mice compared with that in the MHC class-II−/− CD4-Cre Twist2+/+ mice, and there was a significant reduction in the numbers of TCRβ+CD24lo CD8 SP thymocytes in the former mice (Fig. 3d–f and Supplementary Fig. S6a). In addition, analysis of splenocytes revealed that CD8 SP differentiation was severely affected (Supplementary Fig. S6b).
Our results showed that MHC class-II-selected thymocytes were strongly redirected from the CD4+ to the CD8+ T-cell lineage when Twist2 was overexpressed and, in the absence of Twist2, MHC class-I-selected thymocytes displayed reduced CD8 SP cell differentiation. Thus, Twist2 participates in the CD4/CD8 lineage decision by supporting CD8 SP cell differentiation.
Twist2 can regulate ThPOK expression
Many factors known to participate in CD4/CD8 lineage commitment act by modulating ThPOK gene expression. Because Twist2 participates in the CD4/CD8 lineage decision by supporting CD8 SP cell differentiation, we investigated whether Twist2 also regulates CD4/CD8 lineage differentiation via ThPOK.
We found that Twist2 transgenic mice showed significantly reduced ThPOK expression in CD4+CD8lo thymocytes (Supplementary Fig. S7). When we examined ThPOK expression in ThPOK-GFP knock-in mice crossed with Twist2 transgenic mice, the GFP level was significantly reduced in CD4+CD8lo thymocytes from ThPOK-GFP × Twist2 transgenic mice compared with in those from ThPOK-GFP mice (Fig. 4a). While Twist2 transgenic mice showed significantly reduced ThPOK expression in CD4+CD8lo thymocytes, Twist2 cKO mice showed slightly increased ThPOK expression in the same population (Fig. 4b). There was also a slight increase in ThPOK expression in CD8 SP thymocytes (Fig. 4b). These data demonstrate that Twist2 can repress the ThPOK expression.
a ThPOK expression was determined by the relative fluorescence intensity of GFP from the indicated thymocyte subsets from ThPOK-GFP mice and ThPOK-GFP, Twist2 transgenic mice. The numbers above the bracketed lines represent percent GFP-positive cells; numbers in parenthesis below indicate mean fluorescence intensity of GFP in GFP+ cells. Data are from one representative of three independent experiments. b ThPOK expression was determined by the relative fluorescence intensity of GFP from the indicated thymocyte subsets from ThPOK-GFP, CD4-Cre, Twist2+/+ and ThPOK-GFP, CD4-Cre, Twist2f/f mice. The numbers above the bracketed lines represent percent GFP-positive cells; numbers in parenthesis below indicate mean fluorescence intensity of GFP in GFP+ cells. Data are from one representative of three independent experiments. c–d ChIP assay was conducted with CD4 SP, CD8 SP from WT mice or CD4+CD8lo thymocytes from B2m-/-, MHC class-II−/− and MHC class-II−/−, CD4-cre, Twist2f/f mice using (c) anti-H3K4me3 antibody and (d) anti-H3K27me3 antibody. Q-PCR analysis was conducted against ThPOK silencer and P1 promoter regions. (error bars, ±S.E.M., CD4 SP and CD4+CD8lo thymocytes from B2m−/− or MHC class- II−/−: n = 3, CD8 SP: n = 4, CD4+CD8lo thymocytes from MHC class -II−/−, CD4-cre, Twist2f/f: n = 5). *p < 0.05, **p < 0.005, ***p < 0.001 (two-tailed Student’s t test).
It was previously reported that CD8 lineage commitment involves epigenetic ThPOK silencing during a temporal developmental window [22]. Thus, we checked whether Twist2 deficiency could change histone modifications of the ThPOK gene. To analyze epigenetic modifications in the gene locus, we carried out chromatin immunoprecipitation (ChIP) using anti-H3K4me3 and anti-H3K27me3 antibodies. Histone modifications in the ThPOK gene differ depending on whether DP cells become CD4 SP or CD8 SP cells. For CD4 SP cells, the active state (high H3K4me3 levels) was detected in both the silencer and promoter regions (Fig. 4c), whereas repressive H3K27me3 was rarely detected (Fig. 4d). For CD8 SP cells, however, the repressive H3K27me3 was abundant (Fig. 4d), while the active H3K4me3 signal was barely detected (Fig. 4c). To verify whether such differential epigenetic modifications exist at the time of lineage decision, we examined histone modifications of the ThPOK locus in CD4+CD8lo cells from B2m−/− or MHC class-II−/− mice. The CD4+CD8lo cells from B2m−/− mice, which differentiate mostly to CD4 SP cells, showed a high level of H3K4me3 in the silencer and promoter regions of the gene locus, with almost no H3K27me3, as observed with CD4 SP cells. On the other hand, the CD4+CD8lo cells from MHC class-II−/− mice, which differentiate mostly to CD8 SP cells, had a high and low level of the repressive H3K27me3 and active H3K4me3, respectively, in both regions, as observed with CD8 SP cells (Fig. 4c, d). We next evaluated histone modifications of the ThPOK locus in Twist2-deficient MHC class-II−/− mice to see whether the absence of Twist2 induces changes in epigenetic modifications of the ThPOK locus in cells becoming CD8 SP cells (also see Fig. 3e, f). The CD4+CD8lo cells from Twist2-deficient MHC class-II−/− mice showed a high level of H3K27me3 with almost no H3K4me3, similarly to CD8 SP cells or CD4+CD8lo cells of the MHC class-II−/− mice (Fig. 4c, d). These results indicate that epigenetic modifications of the ThPOK gene has already occurred in CD4+CD8lo cells and Twist2 deficiency did not induce any significant change in the epigenetic modifications, resulting in the minor increase in ThPOK expression and, thus, the inefficient generation of CD4 SP cells from MHC class-I-restricted thymocytes (Fig. 3e, f). In addition, another repressor may exist that suppresses the ThPOK expression, even in the absence of Twist2. A recent study showed that Tle3 protein acts as a repressor of ThPOK by binding to the ThPOK silencer region in CD8 SP cells, but not in CD4 SP cells. Therefore, it may be that Tle3 binds to the ThPOK locus, suppressing ThPOK expression in the absence of Twist2. To test this, a ChIP assay was conducted with anti-Tle3 antibody using the CD4+CD8lo cells of MHC class-II−/−, CD4-Cre, Twist2f/f mice. We found that Tle3 is bound to the ThPOK silencer region in the absence of Twist2 (Supplementary Fig. S8). Therefore, both epigenetic modifications and an additional repressor such as Tle appeared to contribute to the inefficient lineage redirection of MHC class-I-restricted thymocytes to CD4 SP cells in Twist2 cKO mice.
Twist2 binds to the silencer region of the ThPOK locus along with the Runx complex
We next examined the mechanisms underlying repressed ThPOK expression during CD8 SP differentiation. Runx3 plays an important role in CD8 SP differentiation by regulating ThPOK expression [20]. It was previously reported that Runx and Twist2 proteins interact [30,31,32]; we also found that Twist2 interacts with Runx3, as assessed by co-immunoprecipitation (Co-IP) analysis (Fig. 5a). Runx complexes (heterodimers of Cbfβ and Runx1 or Runx3) regulate ThPOK expression by binding to the RBS1 and RBS2 regulatory regions in the ThPOK locus [20]. RBS1, a known silencer, is essential for Runx complex-mediated suppression of ThPOK, whereas RBS2 is a known enhancer. Interestingly, Twist2 binds to RBS1 in the ThPOK locus, but not to RBS2 based on ChIP analysis using CD4+CD8lo thymocytes (Fig. 5b, c). Notably, the Twist2:RBS1 interaction was almost completely abrogated in CD4+CD8lo thymocytes from B2m−/− mice, in which CD4 SP differentiation is favored. In contrast, this interaction was strongly enhanced in cells from MHC class-II−/− mice, in which CD8 SP differentiation is favored (Fig. 5c, d). A biotinylated-DNA pull-down assay using biotinylated oligomers corresponding to the NF-κB and E-box region, and Runx-binding region of RBS1 showed that Twist2 was specifically bound to the Runx sites (Fig. 5e, f). Interestingly, Twist2 could bind to the Runx site only if both Twist2 and Runx3 were overexpressed. Almost no binding of Twist2 to the Runx site was observed without concurrent expression of Runx3 (Fig. 5f). Thus, Twist2 appears to repress ThPOK expression by binding to the ThPOK silencer region through interactions with Runx complexes.
a Myc-tagged Runx3 with or without HA-tagged Twist2 was transfected into 293T cells. Whole cell extracts were immunoprecipitated by using anti-Myc antibodies. Western hybridization was conducted by using anti-Myc and anti-HA antibodies. b ChIP assay was conducted with total thymocytes from Twist2 transgenic mice. Q-PCR analysis was conducted against RBS1, RBS2, and UP1 regions. (n = 3) (c, d) CD4+CD8lo thymocytes were sorted from the indicated mice by flow cytometry, followed by ChIP with anti-Twist2 antibodies. c PCR analysis was conducted against the RBS1 and RBS2 region of ThPOK and d Q-PCR analysis was conducted against ThPOK silencer region (error bars, ±S.E.M., n = 3). *p < 0.05 (two-tailed Student’s t test). e Schematic diagram of position of cis-elements and oligomers used for biotinylated-DNA pull-down assay. f Biotinylated-DNA pull-down assay was conducted with biotinylated oligonucleotides, nuclear extracts from 293T cells transfected with HA-Twist2 and myc-Runx3 expressing vectors (upper panel) or only HA-Twist2 expressing vector as a negative control (lower panel), and streptavidin-coated magnetic beads. Pull-downed proteins were analyzed by western hybridization. All experimental data are representative of at least three independent experiments.
Twist2 interacts with the Runx complex in a lineage-specific manner
We investigated the interaction between Twist2 and Runx3 or Cbfβ in distinct thymocyte subsets by using Duolink® analysis. The interaction between Twist2 and Runx3 was dependent on the Twist box domain (Supplementary Fig. S9). Importantly, the Twist2:Runx complex was noted only in CD8 SP thymocytes and no significant interaction was observed in CD8 SP thymocytes from Twist2 cKO mice (Fig. 6a, b). Furthermore, this interaction was more pronounced in CD4+CD8lo thymocytes from MHC class-II−/− mice than in those from B2m−/− mice (Fig. 6c, d), indicating that the Twist2:Runx complex plays distinct roles in the critical step of CD8 lineage differentiation. It has been reported that the Runx complex binds persistently to the RBS1 region of the ThPOK locus in thymocyte subsets, regardless of their capability to differentiate into CD4 or CD8 SP lineages [20]. Thus, it has been postulated that some other factor(s) functions to regulate the distinct suppressive activity of the Runx complex on ThPOK transcription between CD4 and CD8 SP cells. Our results suggest strongly that Twist2 interacts with the Runx complex to suppress ThPOK expression in a CD8 SP lineage-specific manner (Supplementary Fig. S10).
a–d For Duolink in situ PLA analysis, a CD4 SP and CD8 SP cells of WT or Twist2 cKO mice and c CD4+CD8lo thymocyte of WT, B2m−/− and MHC class-II−/− mice were incubated with anti-Twist2 and anti-Cbfβ antibodies, followed by the addition of PLA probes. The fluorescence signal was measured by confocal microscopy. Data are representative of two independent experiments. Scale bars represent 20 μM. Statistical analysis of Duolink data is summarized in b and d. **p < 0.005, ***p < 0.001 (two-tailed Student’s t test).
Discussion
Here, we showed that Twist2 expression is finely regulated during thymocyte differentiation and plays an important role during CD4/CD8 lineage differentiation by controlling ThPOK expression through its interaction with the Runx complex.
We found that the transcription factor Twist2 is tightly regulated during thymocyte development in a differentiation stage-specific manner. Twist2 upregulation was negatively correlated with the intensity of the activation signal, i.e., a weak activation signal for CD8 SP differentiation induced much higher Twist2 expression than a strong signal for CD4 SP differentiation. Consistently, Twist2 expression was differentially regulated when thymocytes differentiated into either CD4 or CD8 SP cells; Twist2 expression was higher in CD8 SP thymocytes than in CD4 SP thymocytes. These results suggest that Twist2 is directly regulated by TCR signaling, which is crucial for thymocyte selection and lineage differentiation processes.
We demonstrated that Twist2 is involved in CD4/CD8 thymocyte lineage determination by regulating a crucial transcription factor, ThPOK, through its interaction with another important factor, the Runx complex. Previous reports indicated that Twist2 interacts with Runx proteins to inhibit their transcriptional activity [30, 31]. In addition, Twist2 expression is associated with the transcriptional downregulation of Runx genes [30]. In our system, Twist2 did not suppress the expression of Runx3 (data not shown). Moreover, like Runx3, Twist2 downregulated the expression of ThPOK both in vitro and in vivo, and the ChIP assay showed that Twist2 binds only to the silencer region among the two major Runx targets. These results suggest that the Twist2:Runx interaction in this system cooperatively suppresses ThPOK expression.
Runx3 binds to the silencer regions of both CD4 and ThPOK to suppress their transcription. As the expression level of CD4 in DP thymocytes can affect the lineage choice, we investigated whether Twist2 can modulate CD4 expression as in the Runx complex, and if constitutive expression of CD4 can block Twist2-induced CD8 SP differentiation. In Twist2 and CD4 double transgenic mice, electronically gated TCRβ+CD24lo cells showed decreases in the CD4 to CD8 SP cell ratio that were similar to the corresponding cells from Twist2 transgenic mice (Supplementary Fig. S11a, b). Therefore, the reduction in mature CD4 SP cells in Twist2 transgenic mice was not likely caused by decreased CD4 expression during DP cell development, but by Twist2 acting directly to drive the CD8 SP cell lineage.
We also showed that Twist2 was associated with altered differentiation of CD4/CD8 thymocyte lineages, but Twist2 deficiency could not efficiently generate lineage-converted MHC class-I-restricted CD4 SP cells in the MHC class-II−/− background. There are a few possible explanations for this inefficient lineage conversion by Twist2 deficiency, which are not necessarily mutually exclusive. One possibility is that epigenetic modification occurred during the CD4+CD8lo stage, and thus the effects of repressor deletion were not able to restore ThPOK expression. Our results showed that the chromatin structure of ThPOK in MHC class-II-restricted cells establishes an active state at the CD4+CD8lo stage. Thus, when repressor proteins such as Twist2 and Runx complexes are overexpressed, they bind to the silencer region of ThPOK to repress its expression, leading to the blockage of CD4 cell differentiation. However, in MHC class-I-restricted cells, chromatin in both the ThPOK silencer and promoter regions appeared to be in a repressive state, and thus re-expression of ThPOK may have been inefficient even in the absence of the repressors. Thus, redirection of differentiation from CD8 T cells to CD4 T cells occurred inefficiently in the Twist2 cKO mice, Runx1 & Runx3 double KO mice, or MAZR KO mice [16, 20]. This process may also require an additional activator, which is absent or expressed at a very low level in CD4+CD8lo thymocytes differentiating into the CD8 lineage (MHC class-II−/− or CD8 SP-derived TCR transgenic mice). Lastly, another repressor(s) for ThPOK suppression may exist and compensate for the absence of Twist2. We found that Tle3 was still bound to the ThPOK silencer region under Twist2-deficient condition, suggesting that Tle-Runx complex still binds to the silencer region and represses the expression of ThPOK. Thus, Tle3, Runx, and Twist2 may form a complex that binds to the ThPOK silencer.
In summary, our results demonstrate that Twist2 is activated by TCR signaling, and its lineage-specific expression pattern and interaction with the Runx complex are involved in regulating ThPOK expression during CD4/CD8 lineage differentiation. Gain-of-function and loss-of-function mutations in Twist2 revealed its critical role in CD8 SP lineage differentiation. The identification of Twist2 as a key player in CD8 T-cell lineage differentiation provides another important clue for understanding the molecular mechanisms underlying the differentiation of thymocytes into CD4/CD8 lineage SP cells.
Methods
Animals
Twist2 transgenic mice and Twist2f/f mice were previously described [27] and detailed mouse generation strategy of Twist2f/f mice is explained in Supplementary Fig. S12. ThPOK-GFP mice have been described [20]. Rag−/− HY-TCR and Rag−/− DO transgenic mice were purchased from Taconic Farms (Korea). Tcra−/− mice and B2m−/− mice were obtained from the Jackson Laboratory. C57BL/6 mice were purchased from Charles River Laboratories. The mice were routinely screened by PCR with DNA obtained from tail tissue. All mouse experiments were approved by the Institutional Animal Care and Use Committee of Seoul National University (No., SNU-151027-5). The mice were bred and maintained under specific pathogen-free conditions, and experiments were performed in accordance with institutional and national guidelines.
Cell lines
293T, HeLa, and 16610D9 cells used in this work have been described before [27, 33]. 293T cells and HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (WelGENE) supplemented with 10% FBS (WelGENE), 100 U/ml streptomycin and penicillin. 16610D9 cells were cultured in Opti-MEM (Invitrogen, Carlsbad, CA, USA) supplemented with 10% FBS, 100 U/ml streptomycin and penicillin.
Antibodies
PerCP-conjugated streptavidin, anti-CD3ε-PE, anti-CD4-PE, anti-CD4-PE-Cy7, anti-CD8-FITC, anti-CD8-APC-Cy7, anti-CD69-PerCP, anti-TCRβ-APC, anti-CD44-FITC, anti-CD25-APC, anti-CD24-PE-Cy7, anti-HY-TCR-APC, and anti-DO11.10-PerCPcy5.5 antibodies were purchased from BD Biosciences or eBioscience. Anti-Twist2 monoclonal antibody (H00117581-M01) and anti-Twist2 polyclonal antibody (ab66031) were purchased from Abnova and abcam, respectively.
Flow cytometry
Stained cells were analyzed or collected on a FACSCanto II or a FACSAria II (BD Biosciences). For cell sorting by surface expression of CD4, CD8, and CD69, a single-cell suspension of thymocytes was stained using anti-CD4-PE, anti-CD8-APC-Cy7, anti-TCRβ-APC, anti-CD24-PE-Cy7, and anti-CD69-PerCP antibodies, and each subset of cells was sorted. The purity of all sorted populations was above 95%.
RT-PCR and Q-PCR
RNA was extracted from the sorted populations or tissues and then used for reverse transcription as described previously [34]. Semi-quantitative RT-PCR was conducted by using a PCR premix kit (ELPIS Biotech, Korea). Quantitative RT-PCR was conducted with primers purchased from Assays-On-Demand (Applied Biosystems) for Twist2, ThPOK, and Runx3 using StepOnePlus® (Applied Biosystems).
ChIP
Total 106 thymocytes were fixed with 1% formaldehyde for 10 min at 37 °C. Cells were collected by centrifugation and washed in cold PBS containing protease inhibitors (1 mM PMSF and protease inhibitor cocktail, Roche) and lysed on ice for 10 min in 200 μl of SDS lysis buffer (Upstate Biotechnology). The chromatin was sheared with an Ultrasonic processor XL (Misonix Inc.). The chromatin fragments are 0.5–1 kilobases in length. After adding tenfold ChIP dilution buffer, sonicated cell supernatants were precleared for 1 h with protein G Sepharose beads and then immunoprecipitated with anti-Twist2 (H00117581-M01) or control mouse IgG1 purified antibodies (Invitrogen, MG100) overnight. Immunoprecipitated complexes were collected on protein G Sepharose beads for 1 h, washed, incubated for 4 h at 65 °C with 5 M NaCl and then for 1 h at 45 °C with proteinase K to reverse the crosslink. DNA was purified by the QiAquick PCR Purification Kit (Qiagen).
Co-IP and western blotting
Myc-tagged Runx3 and/or HA-tagged Twist2 was transfected into 293T cells. Cells were lysed with IPLS lysis buffer (0.05 M Tris-Cl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, and 0.5% NP-40) supplemented with protease inhibitor cocktail (Roche). Cell lysates were immunoprecipitated with anti-Myc antibodies. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to Immobilon-P membrane (Millipore). Blotted proteins were detected using antibodies to Twist2 (ab66031), Myc epitope (9E10,Roche), and HA epitope (HA-7,Sigma) with 5% skim milk in TBST.
Duolink in situ PLA analysis
Transfected HeLa cells or sorted thymocytes were used. The permeabilized HeLa cells/thymocytes were incubated with antibodies against Myc and HA/Twist2 (H00117581-M01; Abnova) and CBFβ (ab33516; Abcam). For secondary antibodies, PLA probes conjugated with a unique short DNA strand (anti-rabbit PLUS and anti-mouse MINUS; Olink Bioscience) were added. After ligation and circularization of the DNA, they were amplified via rolling circle amplification using polymerase and the reactions were detected using a complementary Cy3-labeled DNA linker. The fluorescence signal was detected using confocal microscopy.
Biotinylated-DNA pull-down assay
Four oligonucleotides, containing biotin on the nucleotide at 5′-position, were used in the pull-down assays. The sequences of these oligonucleotides were as follows: Runx-binding site (34mer) F: biotin-5′-GGGGC TGCGG TCTGA GCGCC CCCAG CGGTT TCCT-3′; NF-κB and E box (43mer) F: biotin-5′-GGAGG GGGTA CCCTT GGCAG CCACC GCCTC TTCAG GTGGG TTG-3′; mut Runx-binding site (34mer) F: biotin-5’-GGGGC GAATT CCTGA GCGCC CCCGA ATTCT TCCT-3′; mut NF-κB and E box (43mer) F: biotin-5′-GGAGG TTCTA CCCTT GGCAG CCACC GCCTC TTTAG GTAGG TTG-3′. Each 2 μg of double-stranded oligonucleotide was incubated with 100 μg of nuclear proteins for 20 min at room temperature in a binding buffer containing 20% glycerol, 5 mM MgCl2, 50 mM Tris (pH 7.5), 250 mM NaCl, 2.5 mM EDTA, and 10 μg of poly(dI-dC) competitor. After incubation, 20 μl of streptavidin-coated magnetic beads M-270 (Dynal®, Invitrogen) were added to the reaction and incubated at 4 °C for 4 h. The protein-DNA-streptavidin magnetic bead complex was washed three times with binding buffer and loaded onto a SDS gel. Bounding proteins in the pull-down material were analyzed by Western hybridization using antibodies recognizing HA and Myc-tag.
Statistical analysis
Statistical analysis was performed using Prism 6 (GraphPad). Two-tailed Student t test was used to compare the statistical significance of differences between the samples. The P values are represented in the figures by asterisks (*p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001).
References
von Boehmer H. Selection of the T-cell repertoire: receptor-controlled checkpoints in T-cell development. Adv Immunol. 2004;84:201–38.
Rothenberg EV, Taghon T. Molecular genetics of T cell development. Annu Rev Immunol. 2005;23:601–49.
Bosselut R. CD4/CD8-lineage differentiation in the thymus: from nuclear effectors to membrane signals. Nat Rev Immunol. 2004;4:529–40.
Laky K, Fowlkes BJ. Receptor signals and nuclear events in CD4 and CD8 T cell lineage commitment. Curr Opin Immunol. 2005;17:116–21.
Singer A, Bosselut R. CD4/CD8 coreceptors in thymocyte development, selection, and lineage commitment: analysis of the CD4/CD8 lineage decision. Adv Immunol. 2004;83:91–131.
Kappes DJ, He X, He X. CD4-CD8 lineage commitment: an inside view. Nat Immunol. 2005;6:761–6.
Singer A, Adoro S, Park JH. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat Rev Immunol. 2008;8:788–801.
Mookerjee-Basu J, Chemmannur SV, Qin L, Kappes DJ. ThPOK, a key regulator of T cell development and function. In: Soboloff J, Kappes DJ, editors. Signaling mechanisms regulating T cell diversity and function. Boca Raton (FL): CRC Press/Taylor & Francis; 2018. p. 67–84. https://doi.org/10.1201/9781315371689-5.
Taniuchi I. CD4 helper and CD8 cytotoxic T cell differentiation. Annu Rev Immunol. 2018;36:579–601.
He X, He X, Dave VP, Zhang Y, Hua X, Nicolas E, et al. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–33.
Wilkinson B, Chen JY, Han P, Rufner KM, Goularte OD, Kaye J. TOX: an HMG box protein implicated in the regulation of thymocyte selection. Nat Immunol. 2002;3:272–80.
Woolf E, Xiao C, Fainaru O, Lotem J, Rosen D, Negreanu V, et al. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc Natl Acad Sci USA. 2003;100:7731–6.
Hernandez-Hoyos G, Anderson MK, Wang C, Rothenberg EV, Alberola-Ila J. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity. 2003;19:83–94.
Sun G, Liu X, Mercado P, Jenkinson SR, Kypriotou M, Feigenbaum L, et al. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6:373–81.
Xing S, Shao P, Li F, Zhao X, Seo W, Wheat JC, et al. Tle corepressors are differentially partitioned to instruct CD8(+) T cell lineage choice and identity. J Exp Med. 2018;;215:2211–26.
Sakaguchi S, Hombauer M, Bilic I, Naoe Y, Schebesta A, Taniuchi I, et al. The zinc-finger protein MAZR is part of the transcription factor network that controls the CD4 versus CD8 lineage fate of double-positive thymocytes. Nat Immunol. 2010;11:442–8.
Sakaguchi S, Hainberger D, Tizian C, Tanaka H, Okuda T, Taniuchi I, et al. MAZR and Runx factors synergistically repress ThPOK during CD8+ T cell lineage development. J Immunol. 2015;195:2879–87.
Pai SY, Truitt ML, Ting CN, Leiden JM, Glimcher LH, Ho IC. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 2003;19:863–75.
Nawijn MC, Ferreira R, Dingjan GM, Kahre O, Drabek D, Karis A, et al. Enforced expression of GATA-3 during T cell development inhibits maturation of CD8 single-positive cells and induces thymic lymphoma in transgenic mice. J Immunol. 2001;167:715–23.
Setoguchi R, Tachibana M, Naoe Y, Muroi S, Akiyama K, Tezuka C, et al. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science. 2008;319:822–5.
He X, Park K, Wang H, He X, Zhang Y, Hua X, et al. CD4-CD8 lineage commitment is regulated by a silencer element at the ThPOK transcription-factor locus. Immunity. 2008;28:346–58.
Tanaka H, Naito T, Muroi S, Seo W, Chihara R, Miyamoto C, et al. Epigenetic Thpok silencing limits the time window to choose CD4(+) helper-lineage fate in the thymus. EMBO J. 2013;32:1183–94.
Kohu K, Sato T, Ohno S, Hayashi K, Uchino R, Abe N, et al. Overexpression of the Runx3 transcription factor increases the proportion of mature thymocytes of the CD8 single-positive lineage. J Immunol. 2005;174:2627–36.
Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, et al. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111:621–33.
Li L, Cserjesi P, Olson EN. Dermo-1: a novel twist-related bHLH protein expressed in the developing dermis. Dev Biol. 1995;172:280–92.
Sosic D, Richardson JA, Yu K, Ornitz DM, Olson EN. Twist regulates cytokine gene expression through a negative feedback loop that represses NF-kappaB activity. Cell. 2003;112:169–80.
Oh S, Oh J, Lee C, Oh S, Jeon S, Choi J, et al. Expression of Twist2 is controlled by T-cell receptor signaling and determines the survival and death of thymocytes. Cell Death Differ. 2016;23:1804–14.
Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27.
Kisielow P, Bluthmann H, Staerz UD, Steinmetz M, von Boehmer H. Tolerance in T-cell-receptor transgenic mice involves deletion of nonmature CD4+8+ thymocytes. Nature. 1988;333:742–6.
Sharabi AB, Aldrich M, Sosic D, Olson EN, Friedman AD, Lee SH, et al. Twist-2 controls myeloid lineage development and function. PLoS Biol. 2008;6:e316.
Yang DC, Yang MH, Tsai CC, Huang TF, Chen YH, Hung SC. Hypoxia inhibits osteogenesis in human mesenchymal stem cells through direct regulation of RUNX2 by TWIST. PloS One. 2011;6:e23965.
Pham D, Vincentz JW, Firulli AB, Kaplan MH. Twist1 regulates Ifng expression in Th1 cells by interfering with Runx3 function. J Immunol. 2012;189:832–40.
Davis CB, Killeen N, Crooks ME, Raulet D, Littman DR. Evidence for a stochastic mechanism in the differentiation of mature subsets of T lymphocytes. Cell. 1993;73:237–47.
Lee KY, Choi YI, Kim J, Choi JW, Sohn DH, Lee C, et al. Down-regulation of the SWI/SNF chromatin remodeling activity by TCR signaling is required for proper thymocyte maturation. J Immunol. 2007;178:7088–96.
Acknowledgements
This work was supported by the National Research Foundation of Korea, funded by the Korea government through Grants NRF-2016R1A2B3013865 and Korea Mouse Phenotyping Project Grant NRF-2014M3A9D5A01073789 of the Ministry of Science, ICT and Future Planning through the National Research Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by T. Mak
Supplementary information
Rights and permissions
About this article
Cite this article
Hwang, S., Lee, C., Park, K. et al. Twist2 promotes CD8+ T-cell differentiation by repressing ThPOK expression. Cell Death Differ 27, 3053–3064 (2020). https://doi.org/10.1038/s41418-020-0560-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-020-0560-x