Abstract
Neurodegenerative conditions are characterised by a progressive loss of neurons, which is believed to be initiated by misfolded protein aggregations. During this time period, many physiological and metabolomic alterations and changes in gene expression contribute to the decline in neuronal function. However, these pathological effects have not been fully characterised. In this study, we utilised a metabolomic approach to investigate the metabolic changes occurring in the hippocampus and cortex of mice infected with misfolded prion protein. In order to identify these changes, the samples were analysed by ultrahigh-performance liquid chromatography–tandem mass spectroscopy. The present dataset comprises a total of 498 compounds of known identity, named biochemicals, which have undergone principal component analysis and supervised machine learning. The results generated are consistent with the prion-inoculated mice having significantly altered metabolic profiles. In particular, we highlight the alterations associated with the metabolism of glucose, neuropeptides, fatty acids, l-arginine/nitric oxide and prostaglandins, all of which undergo significant changes during the disease. These data provide possibilities for future studies targeting and investigating specific pathways to better understand the processes involved in neuronal dysfunction in neurodegenerative diseases.
Similar content being viewed by others
Main
Neurodegenerative diseases are characterised by a progressive decline in neuronal health ultimately leading to cell death. Alzheimer’s (AD), Parkinson’s (PD) and Huntington’s diseases are increasingly prevalent neurological conditions that share several common pathways including neuroinflammation, enhanced oxidative stress, metabolic dysfunction and most importantly, the accumulation of misfolded proteins [1,2,3]. One particular form of neuronal degeneration caused by propagating protein misfolding is prion disease, a fatal, progressive neurodegenerative disorder characterised by an accumulation and aggregation of misfolded scrapie-like prion protein PrPSc [4, 5]. Other characteristics of the disease include the development of spongiform lesions, gliosis and neuronal loss, occurring in bovine spongiform encephalopathy (BSE), scrapie, and chronic wasting disease (CWD) in animals and kuru, Creutzfeld–Jacob disease (CJD) and Gerstmann–Sträussler–Scheinker disease in humans [6, 7].
The ubiquitously expressed cellular prion protein (PrPC) is a copper-binding glycosylphosphatidylinositol (GPI)-anchored protein with particularly high expression levels in the brain [8]. It is localised in membrane lipid raft microdomains where it interacts with a variety of proteins involved in regulating synaptic transmission [9, 10]. Prion protein can facilitate synaptic release at glutamatergic synapses [11], is involved in mediating neuronal plasticity by supporting neurite outgrowth [12] and regulates neuronal Ca2+ signalling [13, 14]. PrPC possesses antioxidant activity, thereby reducing the pro-oxidant signalling of free copper ions [15]. During prion disease, the cellular isoform of PrP undergoes a profound conformational rearrangement into a misfolded isoform termed PrPSc that is aggregation-prone and infectious. PrPSc is self-propagating and binding of PrPSc to PrPC leads to the conformational conversion of PrPC into an additional copy of PrPSc [16].
A detailed metabolomic analysis of prion-infected neuronal tissue has not yet been described. In order to generate a snapshot of the different mechanisms that are potentially affected during the development of prion disease, we investigated the metabolic alterations in the cortex and hippocampus of prion-diseased mice. Metabolomics is a powerful approach to monitor and evaluate changes of global metabolic compositions across different biochemical processes caused by changes during disease or other stimulation paradigms [17]. We used mice at 10 weeks post inoculation (wpi) when they present clear characteristics of the disease (e.g. neuronal loss and apoptosis, Fig. 1a–c) but lack confirmatory symptoms, such as weight loss or ataxia (Fig. 1d). Our data demonstrate significant alterations in glucose metabolism, oxidative stress, amino acid utilisation and complex lipid homoeostasis, which can potentially exacerbate progression of the disease. These findings targeting the cortical and hippocampal metabolome present important information for future studies of neurodegeneration.
Prion mice at 10 wpi (RML) exhibit no lack of body weight changes but strong neuronal cell loss. a Neuronal loss in the CA1 hippocampal region is visible in H&E-stained sections of prion-inoculated wpi, see arrow. b TUNEL assay showing apoptotic tissue in RML hippocampus. c NeuN staining of hippocampal CA1 regions from prion-diseased and control (NBH) mice show neuronal loss at 10 wpi in disease. d The weight of prion-inoculated mice remains stable from 6 to 10 wpi, data denote mean ± SEM.
Results
Global metabolic profile in the hippocampus and cortex of prion-diseased mice
We have conducted a thorough metabolomic study of the hippocampus and cortex in prion-infected (RML) mice at 10 wpi and found that both tissues present important biochemical changes compared to age-matched control mice (NBH, see Table 1). The treatment-based separation within the two sample types (prion disease and control) is consistent with the prion disease having a significant impact on metabolically distinct brain regions and pathways. In agreement with these results, a two-way ANOVA analysis identified 141 biochemicals (out of 498) as being significantly affected (p ≤ 0.05) by prion infection (prion main effect). 236 biochemicals, meanwhile, were found to significantly differ within the cortex and hippocampus (region main effect). Overall our data suggests that cellular metabolism is heavily altered by prion disease, with some of the major pathways outlined below (Supplementary Table 1).
Neuropeptide homoeostasis is negatively affected by prion disease
N-acetylaspartate (NAA) is a neuronal precursor of the neurotransmitter N-acetylaspartylglutamate (NAAG, Fig. 2a) that interacts with group II metabotropic glutamate receptors on neurons and glia [18]. Levels of NAA were markedly decreased in cortex and hippocampus of prion-diseased mice (Fig. 2b). Noticeably cortical γ-aminobutyric acid (GABA) levels were also diminished in disease (Fig. 2b) together with a ~30% decrease in GAD 65/67 protein levels (Fig. 2c), an effect not observed in the hippocampus (region main effect p = 0.002). These data suggest an impairment of neuronal function caused by malfunctioning production of neurotransmitter and a subsequent loss of inhibitory GABAergic signalling in the cortex only. Electrophysiological recordings of spontaneous miniature events in the CA1 region showed a 70% decrease in excitatory events, whereas the number of inhibitory events remain unchanged (Fig. 2d) consistent with unchanged hippocampal GABA levels.
Neuropeptide synthesis is downregulated in prion-diseased mice. a Schematic representation of glutamine/glutamate and aspartate metabolism. b Cortex and hippocampus of control and prion-inoculated mice were dissected and levels of indicated metabolites were analysed showing reductions in GABA and NAA levels in prion mice. Enzymes involved in the metabolism are indicated in grey boxes (ABP1 amiloride-binding protein 1, ALDH2 aldehyde dehydrogenase 2, GAD glutamic acid decarboxylase, GLC glutaminase, GS glutamine synthetase, NAAGS N-acetylglutamate synthase, NAT N-acetyltransferase, ASPA/ACY3 aspartate ammonia-lyase, GCPII/III glutamate carboxypeptidase II/III). Data denote mean ± SD and p values are indicated above respective brain regions. c Mean GAD65/67 protein levels in the cortex (left) and hippocampus (right) with representative immunoblots. d Frequency of postsynaptic excitatory and inhibitory currents in CA1 pyramidal neurons shown as number of events with representative recordings. Data denote mean ± SEM
Glucose utilisation is reduced in prion disease
The prion protein plays an important role in glucose metabolism as PrPC has been shown to positively regulate expression of the glucose transporter 1 (Glut 1) with PrPC depletion leading to impaired glucose tolerance [19] and reduced glycolysis [19]. Prion mice present a strong alteration in glucose metabolism characterised by an increased glycolytic flux that translates into an elevated recruitment of the polyol and pentose pathways (Fig. 3a). These mice exhibited a number of changes relating to glucose utilisation. Glucose and several glycolytic pathway intermediates (e.g. fructose 1,6-diphosphate and dihydroxyacetone phosphate [DHAP]), for instance, were present at higher levels in both the hippocampus and cortex samples of the infected animals (compared to control animals). Similar increases were also manifested in metabolites linked to glycogen synthesis and degradation (maltose), polyol synthesis (fructose) and pentose-phosphate-pathway (6-phosphogluconate). An upregulation of the pentose-phosphate-pathway could be explained by an increased demand in NADPH to counteract elevated ROS production. Several other metabolites linked to these pathways were also elevated within the hippocampus alone (Fig. 3a, b). This is consistent with significant changes in glucose availability and utilisation in the prion-infected mice. These changes are particularly relevant given that glucose typically functions as the major fuel source for the brain. Moreover, we found that energy production from malate and pyruvate in RML mice is comparable with that of NBH mice (Fig. 3c), hence an excess of glycolytic metabolites could help maintain ATP production. Evidence suggests that changes in glucose homoeostasis can affect neural function and plasticity [20]. Importantly, the phenotype observed herein could have arisen from several potential factors (e.g. changes in systemic glucose availability and uptake, changes in energetic demand, changes in neural plasticity).
Glucose-related pathways are upregulated in prion-diseased mice. a Schematic representation of the glycolytic, polyol and pentose phosphate pathways. b Cortex and hippocampus of control and prion-inoculated mice were dissected and levels of metabolites analysed. Fructose, maltose and maltotriose levels are increased in diseased hippocampi. Other metabolites are also increased in hippocampal tissues in infected (RML) mice. Data show means ± SD with p values indicated above respective brain regions. c ATP production from malate and pyruvate in the cortex and hippocampus is maintained in prion-diseased mice. Data show means ± SEM
Sphingolipid signalling is modified in prion-inoculated mice
Sphingolipids, or glycosylceramides, are a class of lipids containing a backbone of sphingoid bases and play important roles in signal transmission and cell recognition. Dysfunctional sphingolipid signalling is associated with various diseases, termed sphingolipidoses. Ceramide, a metabolite within this signalling cascade, is the main constituent of the cell membrane and especially in the membranous myelin sheath that surrounds the axons of neurons. Ceramides and sphingosines have pro-apoptotic and antiproliferative properties. Sphingolipid metabolism was altered in the cortex and hippocampus of prion-diseased mice with sphingosine and sphingomyelin (SM) levels both being enhanced in the disease (Fig. 4a, b). We also showed that gene expression of Smpd1, encoding for sphingomyelin phosphodiesterase-1, the enzyme responsible for hydrolysing SM into phosphocholine and ceramide, is significantly downregulated in prion disease (Fig. 4c). Thus, elevated SM levels could be the consequence of reduced break-down in addition to enhanced synthesis, which could represent a compensatory response to advanced neuronal cell death signalling.
Ceramide metabolism is upregulated in prion-diseased mice. a Schematic representation of the membrane phospholipid pathway. b Cortex and hippocampus of control and prion-inoculated mice were dissected and levels of indicated metabolites were analysed showing increased levels of sphingomyelin in prion-infected (RML) mice. c The expression of sphingomyelin phosphodiesterase 1 (Smpd1), encoding for the sphingomyelinase enzyme (SMase), is downregulated in prion-inoculated mice in the hippocampus and cortex, which is responsible for lack of sphingomyelin metabolism. Enzymes involved in ceramide metabolism are indicated in grey boxes: CDase ceramidase, CerS ceramide synthase, SMase sphingomyelinase, SMS sphingomyelin synthase. Data show means ± SD with p values indicated above respective brain regions
Arginine utilisation and nitric oxide (NO) signalling is affected in prion-inoculated mice
l-arginine is a precursor of NO, polyamines, proline, glutamate, creatine and agmatine [21]. It is involved in the nitric oxide synthase (NOS) pathway where l-arginine is converted into NO and l-citrulline and in the arginase pathway. Our data showed that infected animals exhibited a number of changes in arginine metabolism. Most of the arginine-related metabolites were upregulated in prion-inoculated brains (Fig. 5a, b) indicating a strong increase in NO signalling. In addition to the changes outlined above, infected mice also exhibited higher levels of metabolites within the polyamine pathway, such as putrescine and its N-acetylated derivative (N-acetylputrescine) in hippocampal and cortical samples (Fig. 5a, b). These metabolites were the top two metabolites in the random forest (RF) analysis performed with the hippocampus samples (and the 5th and 22nd ranked metabolites in the analysis performed with the cortex samples, see below). They can be derived from the biochemical ornithine in a reaction catalysed by ornithine decarboxylase (ODC) [22]. These alterations are consistent with possible changes in ODC activity (and hence arginine homoeostasis as well, see Fig. 5) and may have further impacts on neural signalling where putrescine can be converted into the GABAergic neurotransmitter.
Altered metabolites of the l-arginine pathway indicate enhanced NO signalling in prion disease. a Schematic representation of arginine indicating an enhanced activity of this pathway. b Cortex and hippocampus of control and prion-inoculated mice were dissected and levels of indicated metabolites were analysed. Data show means ± SD with p values indicated above respective brain regions. c Nitric oxide synthase gene expression (Nos2) is upregulated in disease. Data show means ± SEM with p values indicated above respective brain regions. d NADPH diaphorase staining of a control and prion-inoculated brain shows enhanced signals in prion disease indicating enhanced NOS activity with strong signals in the CA3 and CA1/CA2 hippocampal regions at 10 wpi. Enzymes involved in the metabolism are indicated in grey boxes: ODC ornithine decarboxylase, ARG arginase, iNOS inducible NOS, PRMT protein arginine methyltransferases, DDAH dimethylarginine dimethylaminohydrolase.
All of the above increases are consistent with changes in arginase activity, with concentrations of these compounds typically increasing when there are defects in arginine catabolism. In addition, the infected mice also exhibited significant increases in citrulline (Fig. 5a, b) and trending increases in dimethylarginine (ADMA, Fig. 5a, hippocampus only). These findings closely relate to the enhanced neuroinflammatory activity reported in many neurodegenerative conditions, in which inducible NOS (iNOS) is highly upregulated leading to NO-induced neurotoxicity [23]. This neurotoxic signalling mediates nitrosative stress and can lead to unbalanced formation of NO-mediated post-translational modifications with resulting neuronal and synaptic dysfunction [23,24,25]. The increase in NOS activity in prion mice was confirmed by an elevation of iNOS gene expression (Nos2, Fig. 5c). To specifically examine NOS activity [26] we used the NADPH-diaphorase assay, and found that infected mice showed strong NADPH-diaphorase staining in the hippocampus (Fig. 5d).
Closely associated with enhanced nitrergic stress, our data also support the notion of increased oxidative stress signalling in disease. These changes were mirrored by a reduction in gene expression levels for proteins with antioxidant activity, such as SOD 1 and SOD 2 (Fig. 6a, b), as well as increases in gene expression levels of other pro-oxidant proteins. As such, the robust upregulation of Ncf1, encoding the pore-forming subunit p47phox of the superoxide radical generating NADPH-oxidase enzyme is highly significant in the hippocampus and cortex of diseased mice (Fig. 6a, b), altogether indicating elevated oxidative and nitrergic stress levels. We also found that cytochrome c oxidase (COX IV) protein levels are upregulated by ~80% in the hippocampus (Fig. 6c), which could be a compensatory consequence to maintain respiration levels, oxidative phosphorylation and physiological mitochondrial membrane potential in order to sustain ATP production.
Oxidative stress signalling is enhanced in disease. a Hippocampus, b Cortex. Graphs show relative gene expression of antioxidant and pro-oxidant proteins in both brain regions, SOD 1 and SOD 2 gene expression levels are downregulated in both brain region from diseased mice (RML, red squares) vs. NBH controls (green circles). The pro-oxidant neutrophil cytosol factor 1 (Ncf1) gene expression (NADPH oxidase) is enhanced in prion disease in both brain regions. c COX IV protein levels are upregulated in the hippocampus but not cortex of RML mice. Data show means ± SEM with p values indicated above data sets
Prostaglandin production is upregulated in prion disease
Prostaglandins are lipid compounds involved in inflammation signalling and modulation of neuronal communication. The main axis of prostaglandin signalling is the metabolism of arachidonic acid (AA) via cyclo-oxygenase (COX) 1 and 2 [27]. The gene expression levels for the AA metabolising inducible COX 2 protein were increased in the hippocampus and cortex of prion-infected mice, highlighting the importance of this neuroinflammatory pathway (Fig. 7a, b). In this context, elevated levels of the AA metabolite PGE2 have been found in the cerebrospinal fluid of CJD patients [28]. Our data confirmed that the levels of prostaglandin E2, D2 and F2α were significantly increased in the hippocampus and to a lesser degree in the cortex of prion-diseased mice (Fig. 7c, d). Furthermore, in diseased mice, levels of 12-hydroxyheptadecatrienoic acid (12-HHTrE) were elevated 1.6-fold (p = 3.8 × 10−5) and 1.5-fold (p = 0.044) in the hippocampus and cortex, respectively, providing evidence for strong neuroinflammatory signalling mediated by COX and lipoxygenase pathways (not shown). Supporting the observed increase in neuroinflammation, we found that the macrophage migration inhibitory factor α (Mifα) involved in monocyte recruitment and macrophage inhibition, was increased by ~40% in the hippocampus (Fig. 7e) of RML mice, which also present elevated IκBα gene expression in both cortex and hippocampus (Fig. 7f). IκBα levels strongly correlate with NFκB DNA-binding activity [29], confirming an activation of the anti-inflammatory system in RML mice.
Prostaglandin biosynthesis is upregulated in prion disease. a Schematic representation of prostaglandin biosynthesis. b Gene expression levels for hippocampal (top) and cortical (bottom) COX 2 enzymes. c, d Cortex and hippocampus of control and prion-inoculated mice were dissected and levels of indicated metabolites were analysed. The three main prostaglandins are elevated in the hippocampus of prion infected (RML) mice. Mifα protein levels are increased in the hippocampus of RML mice. e Expression levels of Mifα are slightly elevated in the diseased hippocampus. f Gene expression levels for hippocampal (left) and cortical (right) IκBα. Enzymes involved in the metabolism are indicated in grey boxes: PLA2 phospholipase A II, COX cyclooxygenase, CYP8A prostacyclin synthase, PTGDS prostaglandin-H2 D-isomerase, PTGES prostaglandin-endoperoxide synthase. Data show means ± SD (except (e), (f) with data ± SEM) with p values indicated above respective brain regions
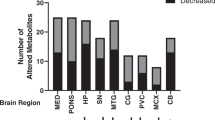
Supervise machine learning analysis of metabolic data
To understand whether measured metabolite concentrations can be used to classify control and prion-inoculated samples we applied RF classification procedure [30]. Estimation of classification accuracy was done using leave-one-out cross-validation [31]. The leave-one-out classification accuracy in the hippocampus vs. control samples was 81% while for the cortex samples vs. control was 87%. Next, we checked the importance of each measured metabolite and its influence on the classification rate. Each metabolite was removed from consideration and leave-one-out cross-validation estimation of classification accuracy repeated on remaining data. The metabolites were ranked next with respect to a decrease in classification accuracy in the cortex (Fig. 8a) and hippocampus (Fig. 8b). For example, exclusion of putrescine from consideration resulted in an almost 33% drop in classification accuracy (~50%) for prion-inoculated hippocampus samples, which is equal to a random classification rate (the same rate is expected if one will assign labels to samples randomly). This finding again indicates the importance of putrescine in the prion-infected hippocampus.
Random forest analysis classifies the levels of importance of indicated metabolites. The importance of each metabolite was estimated by its influence on classification rate (random forest leave-one-out cross-validation). Each metabolite was removed from consideration and leave-one-out cross-validation estimation of classification accuracy repeated on remaining data (see section “Supervise machine learning analysis of metabolic data”). The diagrams plot effect of removing each metabolite on classification accuracy. For example, exclusion of putrescine from consideration resulted in almost 33% drop in classification accuracy (~50%) for prion-inoculated hippocampus samples, which is equal to a random classification rate (the same rate is expected if one will assign labels to samples randomly)
Discussion
Our study characterises metabolic changes occurring at mid-stages during prion disease when memory deficits, neuronal loss and increased apoptosis can be detected (Fig. 1) [32]. We found alterations in metabolic pathways relevant to neurotransmitter signalling, neuroinflammation, oxidative and nitrergic stress energy usage and thus provide data to support research strategies investigating potential pathways for disease rescue [33, 34].
Altered glutamate neuropeptide homoeostasis in disease
Decreased levels of NAA, involved in myelin lipid turnover and neuronal mitochondrial energy production from glutamate [35] are linked to neuronal and axonal loss [36], compromised neuronal metabolism and mitochondrial dysfunction. Reduced NAA levels in AD patient posterior cingulate gyrus and the hippocampus link NAA deficits to attention and memory deficiency [37]. Glutamate carboxypeptidases are zinc-metalloproteins responsible for catalysing the hydrolysis of NAAG to NAA and glutamate [38]. At high concentrations, NAAG has been shown to indirectly inhibit the release of neurotransmitters, such as GABA and glutamate [18]. Thus, a reduction in this arm of glutamatergic neuropeptide metabolism could reflect a compensatory response to reduce excitation and excitotoxicity in order to counter balance reduced inhibitory signalling, although we could not detect changes in glutamate levels. However, our data show that, consistent with unchanged GABA levels in the hippocampus, GAD65/67 protein levels are also unaffected by the disease. This was further substantiated by unchanged inhibitory neuronal activity in pyramidal CA1 neurons, which was in contrast to a decrease in excitatory activity suggesting a neurotransmitter-specific dysfunction in the hippocampus.
Reduced glycolysis and energy production
Data related to the glycolysis, polyol and pentose-phosphate-pathways indicate a strong upregulation of their metabolites, which is related to a reduced glucose metabolism in patients with AD [39] or CJD patients [40, 41]. Moreover, the glycolytic pathway is directly linked to energy production via pyruvate, oxidative phosphorylation and ATP production in mitochondria. These data could feed into the hypothesis that a deficient energy metabolism resulting from defective mitochondrial function and increased oxidative damage may contribute to neuronal dysfunction and degeneration [42]. Thus, our findings are linked with human pathology and provide further detailed information on the abnormalities of the glucose and energy metabolism in this disease.
The strongly activated polyol pathway of glycolysis in prion disease and an accumulation of its metabolites suggest a diversion of glucose metabolism in order to compensate for a failing energy production. Similarly increased NO generation is associated with an increase of polyol synthesis in vascular tissue by upregulation of the rate-limiting enzyme aldose reductase [43] which could thereby link increases in NO signalling with disease associated abnormalities in glycolysis. The other arm of the glucose-related metabolism, the pentose-phosphate pathway, is altered in prion disease with NADPH levels being increased in the hippocampus. Classically, levels of NADPH are elevated in order to prevent oxidative stress and maintain redox balance. In prion-infected mice, upregulation of the glycolytic-polyol-pentose phosphate pathways could reflect an attempt to restore adequate conditions for physiological neuronal function.
Neuroinflammatory NO signalling is augmented
We found the majority of metabolites in the arginine pathways were upregulated in prion-disease indicating increased nitrergic activity, which is consistent with elevated levels of S-nitrosylated proteins found in human prion-diseased post-mortem brain specimens [44]. In the brain, physiological NO signalling contributes to regulation of neuronal excitability [45, 46], neurotransmitter release involved in learning and memory, synaptic plasticity and neuroprotection [47]. Impact of NO signalling on synaptic function is mediated via posttranslational modifications of proteins involved in vesicular release across different cellular systems and species [48,49,50,51] indicating the importance of this conserved signalling route. However, cytotoxic nitrergic signalling is associated with the immune response and plays a key role in regulating inflammatory processes and redox stress, ultimately contributing to neuronal degeneration. This pro-inflammatory NO signalling (mediated via iNOS) will, in addition to abnormally regulated neuronal NOS (nNOS) activity, contribute to nitrergic toxicity. In this context, it has to be considered that the endogenous prion protein possesses a neuroprotective function by promoting S-nitrosylation of NMDAR which in turn minimises excitotoxicity mediated via nNOS activation in a negative feedback loop [52]. Thus, loss of PrPC function per se in prion disease may contribute directly to nitrergic toxicity and further neuronal death.
Furthermore, accumulation of l-arginine metabolites (e.g. guanidino compounds, 2-oxoarginine) may produce neuronal damage which has been reported in argininemia [53, 54]. Reduced activity of arginase may divert arginine metabolism toward the neurotoxic guanidino compounds contributing to the neurotoxicity of this signalling cascade. In mammals, putrescine biosynthesis occurs through the decarboxylation of l-ornithine via the enzyme l-ODC which links the l-arginine pathways (Fig. 5) closely to the metabolic signalling of both polyamines and neurotransmitters (Fig. 2). Polyamines have been detected in both neurons and glia [55] and protect mitochondria against mitochondrial permeability transition [56] acting as potent anti-apoptotic factors. Spermine acts as a free-radical scavenger against ROS [57] to prevent oxidation of mitochondrial transmembrane potential-related proteins including glutathione and other proteins containing sulfhydryl groups. However, other reports favour a pro-apoptotic role of polyamines with levels of ODC/polyamines being changed in AD and PD [58,59,60] leading to a marked intracellular accumulation of amine linked to elevated hydrogen peroxide levels [61, 62]. A toxic elevation of polyamines in neurodegenerative conditions could translate into an excessive activation of NMDA-receptors [63, 64] together with an abnormal activation of apoptotic pathways [9, 65].
Metabolism of sphingolipids is enhanced in disease
Accumulating evidence suggests that sphingolipid metabolism is perturbed early in the development of many neurodegenerative diseases, including AD [66], multiple sclerosis [67] and PD [68]. Several post-mortem studies have found elevated levels of ceramides in brains of AD patients [66, 69] substantiating evidence for neuronal susceptibility to ceramide-induced pathology [70]. Despite accumulating evidence suggesting a pivotal role of sphingolipid dysregulation in the pathogenesis of many brain disorders, few studies have attempted to pharmacologically modulate sphingolipid distribution in the brain. Ceramides have been implicated as a lipid mediator of cellular responses, including redox homoeostasis, inflammation and apoptosis [71]. Forming the central metabolite in the sphingolipid metabolic network, ceramides exist as both ubiquitous modulators of membrane dynamics and as a second messenger [72]. There is a growing body of evidence suggesting increased ceramide facilitates permeabilisation of the mitochondrial membrane (for review see [73]) and potentiates cell death with augmentation in ceramide levels leading to apoptosis and further neuronal degeneration [74, 75]. The reported high levels of ceramide in neural tissue from AD patients [69] indicate this pathway as a strong potential contributor to neurodegeneration. In this context, we found reduced expression of Smpd1 mRNA in prion mice, potentially as a protective mechanism to support myelin repair and oligodendrocyte survival as reported previously [76].
Interestingly, oxidative stress activates sphingomyelinases which in turn hydrolyse SM to yield phosphorylcholine and ceramide, thereby providing a potential link between enhanced ROS signalling and altered ceramide metabolism. A further augmented oxidative cellular environment could result from the lack of cellular prion function as an antioxidant in disease following its misfolding [77] and downregulation of SOD proteins. Thus, it is conceivable to suggest that compromised prion function due to loss of PrPC additionally contributes to the progression of the disease.
Prostaglandin-mediated inflammation is enhanced
As several neurodegenerative conditions are associated with inflammation, our data support the notion that, in addition to inflammatory NO signalling, prostaglandin production could contribute to this pathway. We demonstrated enhanced prostaglandin levels in prion-diseased mice. Indeed, extensive microglial activation induces aggravated inflammatory responses, which contribute to the production of prostaglandins in prion disease [28]. Specifically, upregulation of the enzyme COX 2 (Fig. 7) promotes elevated PGE2 production [78]. Our findings, showing elevated levels of IkBα and by extention enhanced NFκB activation confirm the notion of increased neuroinflammation, However, the effects of prostaglandin signalling are controversial with one report describing PGE2 as downregulating macrophage/microglia activation [79] and indicating prostaglandin-mediated clearance of apoptotic neurons [80]. Another hypothesis favours PGE2 acting as a neurotoxic mediator that facilitates prion-related neuronal apoptosis [81]. Our findings are in agreement with the fact that non-steroidal anti-inflammatory drugs (NSAIDs), which inhibit the metabolism of AA by COX enzymes to eicosanoids, have been implicated as a potential therapeutic target for neurodegenerative disease [82, 83].
Conclusions
This global metabolomic profiling study was conducted to gain insight into mechanisms underlying the impact of prion disease on metabolism in the murine brain. Hence we assessed cortical and hippocampal samples from mice inoculated with misfolded prion protein and healthy age-matched controls. Together, our data provide new information regarding important metabolic pathways which are dysregulated in prion disease. At 10 wpi, neural tissue from prion-inoculated mice has undergone profound changes in metabolism that could indicate an attempt to counteract neuronal degeneration although it is not possible to conclude about the causal relationship between neuronal loss and the observed changes in metabolites. Future studies will have to elucidate at which time points of the disease the reported changes in metabolism occur. These pathways are of great interest for neurodegenerative research as many metabolites identified in this study found to be altered also in AD and PD.
An important outcome of our study is the potential for developing biomarkers or diagnostic tools. Our data provide valuable information for characterising biomarker detection in a non-invasive ante mortem tests or procedures of assessing blood, CSF or urine samples. As such, urine is not subject to homoeostatic mechanisms and accommodate accurate changes that may reflect status of the body, such as in disease and aging [84]. Metabolomics of urine samples can provide certain information to characterise or even predict neurological diseases, including BSE [85, 86]. For instance, the biomarker α1-antichymotrypsin (α1-ACT) is highly upregulated in brains from prion-infected mice and dramatically and progressively increase in urine of patients suffering from sporadic CJD. Elevated levels of α1-ACT were also found in cases of prion disease of animals [87]. Therefore, measurement of urinary metabolites may be useful for monitoring the efficacy of therapeutic regimens for neurodegenerative diseases. Thus, further assessment of metabolic profiles and detailed description of altered pathways will lead to a better understanding of the underlying mechanisms of neurodegeneration and potentially aid in the development of biomarkers for therapeutic targets.
Materials and methods
Mice
All animal work conformed to the UK Home Office regulations and institutional guidelines and subjected to listed procedures under the Project Licence PPL 70/8808 released from the Home Office. All mice were housed under a daily 12 h light/dark cycle. Tg37 mice overexpressing three times the normal levels of murine PrPC were inoculated with either Rocky Mountain Laboratories scrapie prion (RML; n = 19) or normal brain homogenate (NBH; n = 19) at 3 weeks of age. Inoculated mice were then divided into n = 8 and n = 11 per group for metabolomic and mRNA/protein expression/ATP experiments, respectively. All mice were sacrificed at 10 wpi and the hippocampus and cortex were dissected and flash frozen in liquid nitrogen for storage at −80 oC. The time point of 10 wpi was chosen as the animals at this stage show subclinical prion disease without severe signs of neuropathology.
Metabolic profiling
The metabolites were measured by Metabolon Inc. (Durham, NC, USA), a commercial supplier of metabolic analyses. Their platform integrates the chemical analysis, including identification and relative quantification, data reduction, and quality assurance components of the process. Workflow including extraction of raw data, peak-identification, and quality control (QC) processed on the assay platform, has been described [88].
Ultrahigh performance liquid chromatography-tandem mass spectroscopy (UPLC–MS/MS)
All methods utilised a Waters ACQUITY ultra-performance liquid chromatography (UPLC) and a Thermo Scientific Q-Exactive high resolution/accurate mass spectrometer interfaced with a heated electrospray ionisation (HESI-II) source and Orbitrap mass analyser operated at 35,000 mass resolution. The sample extract was dried then reconstituted in solvents compatible to each of the four methods. Each reconstitution solvent contained a series of standards at fixed concentrations to ensure injection and chromatographic consistency. One aliquot was analysed using acidic positive ion conditions, chromatographically optimised for more hydrophilic compounds. In this method, the extract was gradient eluted from a C18 column (Waters UPLC BEH C18-2.1 × 100 mm, 1.7 µm) using water and methanol, containing 0.05% perfluoropentanoic acid (PFPA) and 0.1% formic acid (FA). Another aliquot was also analysed using acidic positive ion conditions, however, it was chromatographically optimised for more hydrophobic compounds. In this method, the extract was gradient eluted from the same afore mentioned C18 column using methanol, acetonitrile, water, 0.05% PFPA and 0.01% FA and was operated at an overall higher organic content. Another aliquot was analysed using basic negative ion optimised conditions using a separate dedicated C18 column. The basic extracts were gradient eluted from the column using methanol and water, however with 6.5 mM ammonium bicarbonate at pH 8. The fourth aliquot was analysed via negative ionisation following elution from a HILIC column (Waters UPLC BEH Amide 2.1 × 150 mm, 1.7 µm) using a gradient consisting of water and acetonitrile with 10 mM ammonium formate, pH 10.8. The MS analysis alternated between MS and data-dependent MSn scans using dynamic exclusion. The scan range varied slighted between methods but covered 70–1000m/z.
The experimental samples were separated by brain region and treatment type when analysed by principal component analysis
Briefly, the experimental samples segregated largely based on brain region (cortex vs. hippocampus) and their treatment status (prion-infected [RML]vs. control [NBH]). The separation between the hippocampal and cortex samples likely reflects several factors (e.g. differences in their functions, cell types and abundance). The treatment-based separation within these two sample types, meanwhile, is consistent with the infection having a significant impact on these metabolically distinct brain regions (Supplementary Table 1). In agreement with these results, a two-way ANOVA analysis identified 141 biochemicals as being significantly affected (p ≤ 0.05) by prion infection (infection main affect). 236 biochemicals, meanwhile, were found to significantly differ within the cortex and hippocampus (region main effect). It should be noted however, that there were only 24 biochemicals displaying significant interaction effects (i.e. these correspond to biochemicals which changed in differential manners consequent to infection in the hippocampus and cortex) with the full list of detected metabolites shown in Supplementary Table 1. The metabolic changes arising from the infection appeared to be highly conserved therefore in the two brain regions.
RF analysis distinguished between the cortex and hippocampal samples of the control and prion-inoculated groups
RF is an unbiased and supervised classification technique based on an ensemble of a large number of decision trees. In addition to producing a metric of predictive accuracy, RF analysis also produces an associated list of biochemicals ranked in order of their importance to the classification scheme. In this study, RF analysis was used to identify metabolites that differentiated between the cortex and hippocampal samples derived from the control (NBH) and prion-infected (RML) mice. The predictive accuracies of these analyses were 87% and 81%, respectively. Like the PCA analysis, these results are consistent with the prion-infected mice having significantly altered metabolic profiles.
mRNA expression
Total RNA was extracted from isolated hippocampal or cortical tissue using a RNeasy mini kit and treated with deoxyribonuclease 1. For each tissue, 1 µg of total RNA was reverse transcribed using an iScript™ cDNA synthesis kit according to the manufacturer’s instructions. Real-time PCR reactions were performed using 25–100 ng of template cDNA per reaction and Taqman gene expression ‘assay on demand’ assays. The primer/probe targets used were FAM –labelled Ncf1 (Mm00447921_m1), Nos2 (Mm00440502_m1), Smpd1 (Mm00488318_m1), Sod1 (Mm01700393_g1), Sod2 (Mm01313000_m1), Nfkbia (Mm00477798_ml) and Ptgs2 (Mm00478374_m1). Reactions were multiplexed and normalised to a VIC-labelled primer/probe for β-actin (Actb; Mm00607939_s1). Relative mRNA expression in each region was determined by normalising to the average of NBH-treated hippocampus or cortex using the ΔΔCT method.
NADPH-diaphorase staining
Mice were anaesthetised and transcardially perfused with PBS then 4% PFA. The brain was removed and fixed for 24 h in PFA before being embedded in paraffin. Five μm sections were cut and deparaffinised before being incubated in a mixture of 2.5 mg nitroblue tetrazolium, 10 mg β-NADPH, and 0.2% Triton X-100 in 10 mL of 0.05 M Tris buffer for 30 min at room temperature. The reaction was terminated with 0.05 M Tris buffer washes.
Electrophysiology
Whole cell recordings were made from CA1 pyramidal neurons using a Multiclamp 700B amplifier and pClamp 10.4 software (Molecular Devices), sampling at 50 kHz and filtering at 10 kHz. Patch pipettes were pulled from filamented borosilicate glass (GC150F-7.5, outer diameter 1.5 mm; inner diameter 0.86 mm; Harvard Apparatus, Edenbridge, UK) with a two-stage vertical puller (PC-10 Narishige, Tokyo, Japan). Pipettes were used with final tip resistance of 4–5 MΩ when filled with a solution containing (mM): 120 mM potassium methanesulfonate, HEPES 10, EGTA 0.2,K-ATP 4, Na-GTP 0.3, NaCl 8, KCl 10; pH was adjusted to 7.4 with KOH and osmaolarity between 280–290 mosmol l−1. All recordings were taken at physiological temperature of 37 ± 1 °C, with the bath temperature being feedback controlled by a Peltier device warming aCSF passing through a fast-flow perfusion system. Inhibitory and excitatory miniature postsynaptic currents were analysed within 50 s of recordings at a holding potential of −60 mV.
TUNEL Assay
Following PFA fixation, brains were processed in paraffin wax and sliced at 5 μM using a microtome. TUNEL assay was performed as per manufacturer’s recommendations (In situ apoptosis detection kit, ab206386) on paraffin-fixed brain sections.
ATP production
Hippocampus and cortex dissected from 8 mice (NBH = 4 and RML = 4) were homogenised in 10 volume ice-cold TES buffer (10 mM HPO42−/H2PO4−, 2 mM K-EDTA, 250 mM sucrose, pH 7.4) by disrupting the tissue using a glass homogeniser. The samples were then centrifuged (600×g, 4 oC, 15 min) and the supernatant recovered. Following the Bradford assay, samples of equivalent protein concentration were incubated with the reaction buffer (30 mM HPO42−/H2PO4−, 75 mM KCl, 0.5 mM MgCl2, 1 mM K-EDTA, 2 mM ADP, 1 mM pyruvate, 1 mM malate, 20 mM creatine and 0.2 mM P1, P5-di-adenosine 5′-pentaphosphate, pH 7.4) for 5, 10, 15, 20, and 30 min. Reactions were terminated by immediately transferring the samples on ice. After centrifugation (14,000×g, 4 oC, 15 min), the ATP luciferase assay was performed as per manufacturer’s instructions (A22066, Molecular Probes) in duplicate. Luminescence was quantified with a CLARIOstar.
Immunoblotting
Mice were culled via cervical dislocation and hippocampi and cortex were dissected and snap frozen on dry ice. Protein samples were isolated using protein lysis buffer (50 mM Tris–HCl, 1 mM EDTA, 1 mM EGTA, 0.1% β-mercaptoethanol, 1% triton X-100, pH 7.5) supplemented with protease inhibitors (Roche). Concentrations of proteins in the lysate were determined using a Bradford assay. 5 μg of protein were mixed with loading buffer and boiled for 5 min before loading onto a 12% SDS–polyacrylamide electrophoresis gels and transferred onto nitrocellulose membrane, using a Bio-Rad semi-dry transfer system. Membranes were blocked in 5% milk/TBST for 1 h at room temperature. Membranes were incubated overnight at 4 °C with anti-GAD 65/67 (AB1511), anti-COX IV (ab14744), anti-Mifα (HPA003868) or anti-β-actin (ab8227) antibodies followed by incubation with appropriate secondary antibodies.
Immunocytochemistry
Following PFA fixation, brains were processed in paraffin wax and sliced at 5 μM using a microtome. After antigen retrieval, sections were washed in TBS + 0.1% triton x-100 and blocked for 2 h at RT in TBS, 0.1% triton X-100, 10% goat serum and 5% BSA. Sections were incubated with antibodies to NeuN (ab104225) at 1:1000 in blocking buffer overnight at 4 °C. Sections were washed three times, and incubated with Alexa Fluor fluorescent secondary antibody for 1 h at RT in blocking buffer. Images were taken with a Zeiss LSM 510 META NLO microscope with Zen 2009 software (Zeiss).
Statistical analysis
Data were analysed using GraphPad Prism 6 and results expressed as mean ± SD/SEM (as indicated), with p < 0.05 considered significant. Two-way ANOVA were performed followed by Welch’s two-sample t-tests to identify significant differences between experimental groups.
References
Villar-Pique A, Schmitz M, Candelise N, Ventura S, Llorens F, Zerr I. Molecular and clinical aspects of protein aggregation assays in neurodegenerative diseases. Mol Neurobiol. https://doi.org/10.1007/s12035-018-0926-y 2018.
Angelova PR, Abramov AY. Role of mitochondrial ROS in the brain: from physiology to neurodegeneration. FEBS Lett. 2018;592:692–702.
Neal M, Richardson JR. Epigenetic regulation of astrocyte function in neuroinflammation and neurodegeneration. Biochim Biophys Acta. 2018;1864:432–43.
Aguzzi A, Heppner FL. Pathogenesis of prion diseases: a progress report. Cell Death Differ. 2000;7:889–902.
Watts JC, Prusiner SB. Mouse models for studying the formation and propagation of prions. J Biol Chem. 2014;289:19841–9.
Parchi P, Strammiello R, Giese A, Kretzschmar H. Phenotypic variability of sporadic human prion disease and its molecular basis: past, present, and future. Acta Neuropathol. 2011;12:91–112.
Scheckel C, Aguzzi A. Prions, prionoids and protein misfolding disorders. Nat Rev Genet. https://doi.org/10.1038/s41576-018-0011-4 2018.
Bendheim PE, Brown HR, Rudelli RD, Scala LJ, Goller NL, Wen GY, et al. Nearly ubiquitous tissue distribution of the scrapie agent precursor protein. Neurology. 1992;42:149–56.
Westergard L, Christensen HM, Harris DA. The cellular prion protein (PrP(C)): its physiological function and role in disease. Biochim Biophys Acta. 2007;1772:629–44.
Steinert JR. Prion protein as a mediator of synaptic transmission. Commun Integr Biol. 2015;8:e1063753.
Robinson SW, Nugent ML, Dinsdale D, Steinert JR. Prion protein facilitates synaptic vesicle release by enhancing release probability. Hum Mol Genet. 2014;23:4581–96.
Chen S, Mangé A, Dong L, Lehmann S, Schachner M. Prion protein as trans-interacting partner for neurons is involved in neurite outgrowth and neuronal survival. Mol Cell Neurosci. 2003;22:227–33.
Krebs B, Wiebelitz A, Balitzki-Korte B, Vassallo N, Paluch S, Mitteregger G, et al. Cellular prion protein modulates the intracellular calcium response to hydrogen peroxide. J Neurochem. 2007;100:358–67.
Fuhrmann M, Bittner T, Mitteregger G, Haider N, Moosmang S, Kretzschmar H, et al. Loss of the cellular prion protein affects the Ca2+ homeostasis in hippocampal CA1 neurons. J Neurochem. 2006;98:1876–85.
Brown DR. Neurodegeneration and oxidative stress: prion disease results from loss of antioxidant defence. Folia Neuropathol/Assoc Pol Neuropathol Med Res Cent, Pol Acad Sci. 2005;43:229–43.
Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–44.
Fiehn O. Combining genomics, metabolome analysis, and biochemical modelling to understand metabolic networks. Comp Funct Genom. 2001;2:155–68.
Zhou J, Neale JH, Pomper MG, Kozikowski AP. NAAG peptidase inhibitors and their potential for diagnosis and therapy. Nat Rev Drug Discov. 2005;4:1015–26.
Li Q-Q, Sun Y-P, Ruan C-P, Xu X-Y, Ge J-H, He J, et al. Cellular prion protein promotes glucose uptake through the Fyn-HIF-2α-Glut1 pathway to support colorectal cancer cell survival. Cancer Sci. 2011;102:400–6.
Mainardi M, Fusco S, Grassi C. Modulation of hippocampal neural plasticity by glucose-related signaling. Neural Plast. 2015;2015:657928.
Morris SM. Arginine metabolism: boundaries of our knowledge. J Nutr. 2007;137:1602S–9S.
Babu GN, Sailor KA, Beck J, Sun D, Dempsey RJ. Ornithine decarboxylase activity in in vivo and in vitro models of cerebral ischemia. Neurochem Res. 2003;28:1851–7.
Steinert JR, Chernova T, Forsythe ID. Nitric oxide signaling in brain function, dysfunction, and dementia. Neuroscience. 2010;16:435–52.
Nakamura T, Lipton SA. S-Nitrosylation and uncompetitive/fast off-rate (UFO) drug therapy in neurodegenerative disorders of protein misfolding. Cell Death Differ. 2007;14:1305–14.
Bradley SA, Steinert JR. Nitric oxide-mediated posttranslational modifications: impacts at the synapse. Oxid Med Cell Longev. 2016;2016:5681036.
Hope BT, Michael GJ, Knigge KM, Vincent SR. Neuronal NADPH diaphorase is a nitric oxide synthase. Proc Natl Acad Sci. 1991;88:2811–4.
Helliwell RJ, Adams LF, Mitchell MD. Prostaglandin synthases: recent developments and a novel hypothesis. Prostaglandins Leukot Essent Fat Acids. 2004;70:101–13.
Minghetti L, Pocchiari M. Cyclooxygenase-2, prostaglandin E2, and microglial activation in prion diseases. Int Rev Neurobiol. 2007;82:265–75.
Bottero V, Imbert V, Frelin C, Formento JL, Peyron JF. Monitoring NF-kappa B transactivation potential via real-time PCR quantification of I kappa B-alpha gene expression. Mol Diagn. 2003;7:187–94.
Cutler DR, Edwards TC Jr., Beard KH, Cutler A, Hess KT, Gibson J, et al. Random forests for classification in ecology. Ecology. 2007;88:2783–92.
Vapnik VN. An overview of statistical learning theory. IEEE Trans Neural Netw. 1999;10:988–99.
Moreno JA, Radford H, Peretti D, Steinert JR, Verity N, Martin MG, et al. Sustained translational repression by eIF2alpha-P mediates prion neurodegeneration. Nature. 2012;485:507–11.
Peretti D, Bastide A, Radford H, Verity N, Molloy C, Martin MG, et al. RBM3 mediates structural plasticity and protective effects of cooling in neurodegeneration. Nature. 2015;518:236–9.
Bradley SJ, Bourgognon JM, Sanger HE, Verity N, Mogg AJ, White DJ, et al. M1 muscarinic allosteric modulators slow prion neurodegeneration and restore memory loss. J Clin Invest. 2017;127:487–99.
Chakraborty G, Mekala P, Yahya D, Wu G, Ledeen RW. Intraneuronal N-acetylaspartate supplies acetyl groups for myelin lipid synthesis: evidence for myelin-associated aspartoacylase. J Neurochem. 2001;78:736–45.
Demougeot C, Garnier P, Mossiat C, Bertrand N, Giroud M, Beley A, et al. N-Acetylaspartate, a marker of both cellular dysfunction and neuronal loss: Its relevance to studies of acute brain injury. J Neurochem. 2001;77:408–15.
Schuff N, Capizzano AA, Du AT, Amend DL, O’Neill J, Norman D, et al. Selective reduction of N-acetylaspartate in medial temporal and parietal lobes in AD. Neurology. 2002;58:928–35.
Rojas C, Frazier ST, Flanary J, Slusher BS. Kinetics and inhibition of glutamate carboxypeptidase II using a microplate assay. Anal Biochem. 2002;310:50–4.
Willette AA, Bendlin BB, Starks EJ, Birdsill AC, Johnson SC, Christian BT, et al. Association of insulin resistance with cerebral glucose uptake in late middle–aged adults at risk for alzheimer disease. JAMA Neurol. 2015;72:1013–20.
Kim EJ, Cho SS, Jeong BH, Kim YS, Seo SW, Na DL, et al. Glucose metabolism in sporadic Creutzfeldt-Jakob disease: a statistical parametric mapping analysis of (18) F-FDG PET. Eur J Neurol. 2012;19:488–93.
Renard D, Vandenberghe R, Collombier L, Kotzki PO, Pouget JP, Boudousq V. Glucose metabolism in nine patients with probable sporadic Creutzfeldt-Jakob disease: FDG-PET study using SPM and individual patient analysis. J Neurol. 2013;260:3055–64.
Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–95.
Seo HG, Nishinaka T, Yabe-Nishimura C. Nitric oxide up-regulates aldose reductase expression in rat vascular smooth muscle cells: a potential role for aldose reductase in vascular remodeling. Mol Pharmacol. 2000;57:709–17.
Chen LN, Shi Q, Zhang BY, Zhang XM, Wang J, Xiao K, et al. Proteomic analyses for the global S-nitrosylated proteins in the brain tissues of different human prion diseases. Mol Neurobiol. 2016;53:5079–96.
Steinert JR, Robinson SW, Tong H, Haustein MD, Kopp-Scheinpflug C, Forsythe ID. Nitric oxide is an activity-dependent regulator of target neuron intrinsic excitability. Neuron. 2011;71:291–305.
Steinert JR, Kopp-Scheinpflug C, Baker C, Challiss RA, Mistry R, Haustein MD, et al. Nitric oxide is a volume transmitter regulating postsynaptic excitability at a glutamatergic synapse. Neuron. 2008;60:642–56.
Hardingham N, Dachtler J, Fox K. The role of nitric oxide in pre-synaptic plasticity and homeostasis. Front Cell Neurosci. 2013;7:190.
Wiseman DA, Kalwat MA, Thurmond DC. Stimulus-induced S-nitrosylation of Syntaxin 4 impacts insulin granule exocytosis. J Biol Chem. 2011;286:16344–54.
Palmer ZJ, Duncan RR, Johnson JR, Lian LY, Mello LV, Booth D, et al. S-nitrosylation of syntaxin 1 at Cys(145) is a regulatory switch controlling Munc18-1 binding. Biochem J. 2008;413:479–91.
Robinson SW, Bourgognon JM, Spiers JG, Breda C, Campesan S, Butcher A, et al. Nitric oxide-mediated posttranslational modifications control neurotransmitter release by modulating complexin farnesylation and enhancing its clamping ability. PLoS Biol. 2018;16:e2003611.
Wang G, Moniri NH, Ozawa K, Stamler JS, Daaka Y. Nitric oxide regulates endocytosis by S-nitrosylation of dynamin. Proc Natl Acad Sci USA. 2006;103:1295–300.
Gasperini L, Meneghetti E, Pastore B, Benetti F, Legname G. Prion protein and copper cooperatively protect neurons by modulating NMDA receptor through S-nitrosylation. Antioxid Redox Signal. 2015;22:772–84.
Wiechert P, Mortelmans J, Lavinha F, Clara R, Terheggen HG, Lowenthal A. Excretion of guanidino-derivates in urine of hyperargininemic patients. J Genet Hum. 1976;24:61–72.
Deignan JL, De Deyn PP, Cederbaum SD, Fuchshuber A, Roth B, Gsell W, et al. Guanidino compound levels in blood, cerebrospinal fluid, and post-mortem brain material of patients with argininemia. Mol Genet Metab. 2010;100:S31–6.
Bernstein H-G, Müller M. The cellular localization of the l-ornithine decarboxylase/polyamine system in normal and diseased central nervous systems. Prog Neurobiol. 1999;57:485–505.
Marcocci L, Marchi U, Salvi M, Milella ZG, Nocera S, Agostinelli E, et al. Tyramine and monoamine oxidase inhibitors as modulators of the mitochondrial membrane permeability transition. J Membr Biol. 2002;188:23–31.
Sava IG, Battaglia V, Rossi CA, Salvi M, Toninello A. Free radical scavenging action of the natural polyamine spermine in rat liver mitochondria. Free Radic Biol Med. 2006;41:1272–81.
Morrison LD, Kish SJ. Brain polyamine levels are altered in Alzheimer’s disease. Neurosci Lett. 1995;197:5–8.
Morrison LD, Cao X-C, Kish SJ. Ornithine decarboxylase in human brain: influence of aging, regional distribution, and Alzheimer’s disease. J Neurochem. 1998;71:288–94.
Gomes-Trolin C, Nygren I, Aquilonius SM, Askmark H. Increased red blood cell polyamines in ALS and Parkinson’s disease. Exp Neurol. 2002;177:515–20.
Toninello A, Mondovì MS. Interaction of biologically active amines with mitochondria and their role in the mitochondrial-mediated pathway of apoptosis. Curr Med Chem. 2004;11:2349–74.
Stefanelli C, Bonavita F, Stanic’ I, Pignatti C, Flamigni F, Guarnieri C, et al. Spermine triggers the activation of caspase-3 in a cell-free model of apoptosis. FEBS Lett. 1999;451:95–8.
Sparapani M, Dall’Olio R, Gandolfi O, Ciani E, Contestabile A. Neurotoxicity of polyamines and pharmacological neuroprotection in cultures of rat cerebellar granule cells. Exp Neurol. 1997;148:157–66.
Williams K. Modulation and block of ion channels: a new biology of polyamines. Cell Signal. 1997;9:1–13.
Schipper RG, Penning LC, Verhofstad AAJ. Involvement of polyamines in apoptosis. Facts and controversies: effectors or protectors? Semin Cancer Biol. 2000;10:55–68.
Cutler RG, Kelly J, Storie K, Pedersen WA, Tammara A, Hatanpaa K, et al. Involvement of oxidative stress-induced abnormalities in ceramide and cholesterol metabolism in brain aging and Alzheimer’s disease. Proc Natl Acad Sci USA. 2004;101:2070–5.
Vidaurre OG, Haines JD, Katz Sand I, Adula KP, Huynh JL, McGraw CA, et al. Cerebrospinal fluid ceramides from patients with multiple sclerosis impair neuronal bioenergetics. Brain. 2014;137:2271–86.
Xing Y, Tang Y, Zhao L, Wang Q, Qin W, Ji X, et al. Associations between plasma ceramides and cognitive and neuropsychiatric manifestations in Parkinson’s disease dementia. J Neurol Sci. 2016;370:82–7.
Filippov V, Song MA, Zhang K, Vinters HV, Tung S, Kirsch WM, et al. Increased ceramide in brains with Alzheimer’s and other neurodegenerative diseases. J Alzheimer’s Dis. 2012;29:537–47.
Mencarelli C, Martinez-Martinez P. Ceramide function in the brain: when a slight tilt is enough. Cell Mol life Sci. 2013;70:181–203.
Hannun YA. Functions of ceramide in coordinating cellular responses to stress. Science. 1996;274:1855–9.
van Blitterswijk WJ, van der Luit AH, Veldman RJ, Verheij M, Borst J. Ceramide: second messenger or modulator of membrane structure and dynamics? Biochem J. 2003;369:199–211.
Taha TA, Mullen TD, Obeid LM. A house divided: ceramide, sphingosine, and sphingosine-1-phosphate in programmed cell death. Biochim Biophys Acta. 2006;1758:2027–36.
Novgorodov SA, Szulc ZM, Luberto C, Jones JA, Bielawski J, Bielawska A, et al. Positively charged ceramide is a potent inducer of mitochondrial permeabilization. J Biol Chem. 2005;280:16096–105.
Obeid LM, Linardic CM, Karolak LA, Hannun YA. Programmed cell death induced by ceramide. Science. 1993;259:1769 LP–71.
Chami M, Halmer R, Schnoeder L, Anne Becker K, Meier C, Fassbender K, et al. Acid sphingomyelinase deficiency enhances myelin repair after acute and chronic demyelination. PLoS One. 2017;12:e0178622–e.
Onodera T, Sakudo A, Tsubone H, Itohara S. Review of studies that have used knockout mice to assess normal function of prion protein under immunological or pathophysiological stress. Microbiol Immunol. 2014;58:361–74.
Walsh DT, Perry VH, Minghetti L. Cyclooxygenase-2 is highly expressed in microglial-like cells in a murine model of prion disease. Glia. 2000;29:392–6.
Harris SG, Padilla J, Koumas L, Ray D, Phipps RP. Prostaglandins as modulators of immunity. Trends Immunol. 2002;23:144–50.
Gray F, Chretien F, Adle-Biassette H, Dorandeu A, Ereau T, Delisle MB, et al. Neuronal apoptosis in Creutzfeldt-Jakob disease. J Neuropathol Exp Neurol. 1999;58:321–8.
Bate C, Rutherford S, Gravenor M, Reid S, Williams A. Cyclo-oxygenase inhibitors protect against prion-induced neurotoxicity in vitro. Neuroreport. 2002;13:1933–8.
Giovannini MG, Scali C, Prosperi C, Bellucci A, Pepeu G, Casamenti F. Experimental brain inflammation and neurodegeneration as model of Alzheimer’s disease: protective effects of selective COX-2 inhibitors. Int J Immunopathol Pharmacol. 2003;16:31–40.
Trepanier CH, Milgram NW. Neuroinflammation in Alzheimer’s disease: are NSAIDs and selective COX-2 inhibitors the next line of therapy? J Alzheimer’s Dis. 2010;21:1089–99.
Wu J, Gao Y. Physiological conditions can be reflected in human urine proteome and metabolome. Expert Rev Proteom. 2015;12:623–36.
An M, Gao Y. Urinary biomarkers of brain diseases. Genom Proteom Bioinforma. 2015;13:345–54.
Plews M, Lamoureux L, Simon SL, Graham C, Ruddat V, Czub S, et al. Factors affecting the accuracy of urine-based biomarkers of BSE. Proteome Sci. 2011;9:6.
Miele G, Seeger H, Marino D, Eberhard R, Heikenwalder M, Stoeck K, et al. Urinary alpha1-antichymotrypsin: a biomarker of prion infection. PLoS One. 2008;3:e3870.
Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem. 2009;81:6656–67.
Acknowledgements
J.B. was funded by The Henry Smith Charity, UK; J.G.S., H.S., A.A. and J.R.S. were supported by the Medical Research Council, UK. This work was further supported in part by a Wellcome Trust Collaborative Award [201529/Z/16/Z] (ABT), a Glasgow University Lord Kelvin Adma Smith Award (SJB) and a Medical Research Council MICA award [MR/P019366/1] (SJB, ABT).
Author Contributions:
J.B., J.G.S., A.A., S.J.B. and H.S. performed experiments and statistical analysis, J.B. and J.R.S. wrote the manuscript and revised the manuscript together with J.G.S. and A.B.T.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Edited by: G. Melino
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Bourgognon, JM., Spiers, J.G., Scheiblich, H. et al. Alterations in neuronal metabolism contribute to the pathogenesis of prion disease. Cell Death Differ 25, 1408–1425 (2018). https://doi.org/10.1038/s41418-018-0148-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-018-0148-x
This article is cited by
-
Quaternary Structure Changes for PrPSc Predate PrPC Downregulation and Neuronal Death During Progression of Experimental Scrapie Disease
Molecular Neurobiology (2021)
-
Metabolomic study of disease progression in scrapie prion infected mice; validation of a novel method for brain metabolite extraction
Metabolomics (2020)
-
Brain energy rescue: an emerging therapeutic concept for neurodegenerative disorders of ageing
Nature Reviews Drug Discovery (2020)