Abstract
Background
A significant subset of prostate cancer (PC) patients with a castration-resistant form of the disease (CRPC) show primary resistance to androgen receptor (AR)-targeting drugs developed against CRPC. As one explanation could be the expression of constitutively active androgen receptor splice variants (AR-Vs), our current objectives were to study AR-Vs and other AR aberrations to better understand the emergence of CRPC.
Methods
We analysed specimens from different stages of prostate cancer by next-generation sequencing and immunohistochemistry.
Results
AR mutations and copy number variations were detected only in CRPC specimens. Genomic structural rearrangements of AR were observed in 5/30 metastatic CRPC patients, but they were not associated with expression of previously known AR-Vs. The predominant AR-Vs detected were AR-V3, AR-V7 and AR-V9, with the expression levels being significantly higher in CRPC cases compared to prostatectomy samples. Out of 25 CRPC metastases that expressed any AR variant, 17 cases harboured expression of all three of these AR-Vs. AR-V7 protein expression was highly heterogeneous and higher in CRPC compared to hormone-naïve tumours.
Conclusions
AR-V3, AR-V7 and AR-V9 are co-expressed in CRPC metastases highlighting the fact that inhibiting AR function via regions common to all AR-Vs is likely to provide additional benefit to patients with CRPC.
Similar content being viewed by others
Introduction
Prostate cancer (PC) is the most common malignancy and third most common cause of cancer-related death among men in Europe. Androgens are required for the normal development of prostate tissue and exert their effects through androgen receptor (AR)-mediated signalling, but also have important role during PC emergence and progression. Most PCs grow slowly and are curable by surgery and radiation when confined to the prostate. In contrast, treatment of PCs that have spread outside the prostate usually includes manipulation of the AR signalling axis; androgen-deprivation therapy (ADT) either by surgical or chemical castration. However, during currently available ADT, lethal castration-resistant form of PC (CRPC) will eventually emerge after a variable period of time. Even though the exact mechanism by which CRPC develops remains to be fully understood, several mechanisms of castration resistance have been identified such as AR gene amplification,1,2 point mutations in AR gene3,4 and induction of steroidogenesis in CRPC cells.5,6,7 AR gene amplification has been demonstrated in approximately 30% of CRPCs.1 Cancers with AR amplification have been shown to respond better to second-line maximal androgen blockade compared to tumours without the amplification, however, the response was short-lived.8 Also AR mutations are rare even at CRPC stage being present in approximately 10–30% of cases.4 These mutations are almost always associated with diverse gains-of-function and about 45% of the mutations occur in the ligand-binding domain.9 AR mutations can broaden ligand specificity to alternative steroid hormones, hypersensitise the receptor to castrate levels of androgens or lead to resistance to current forms of treatment making AR active even in the presence of anti-androgens.10
Importantly, androgen signalling remains active even in the CRPC stage.11,12 The established concept of sustained AR signalling during CRPC has led to the clinical development of second-generation AR-targeting drugs enzalutamide and abiraterone that target the ligand-binding domain of AR directly and indirectly, respectively. Enzalutamide is an AR antagonist, whereas abiraterone is a CYP17 inhibitor approved by the US Food and Drug Administration (FDA) for the treatment of metastatic CRPC. Several studies have shown that presence of AR amplification or AR mutations in plasma samples is associated with worse outcome with enzalutamide and abiraterone.13,14,15,16,17 Furthermore, a significant subset of patients show primary resistance to these agents with respect to PSA (prostate-specific antigen) level,18 and among patients who initially respond, nearly all eventually develop acquired resistance.
One potential explanation for the resistance to first-generation and second-generation AR-targeted therapies is the presence of AR splice variants (AR-Vs). AR-Vs are alternatively spliced isoforms of the AR mRNA usually resulting in truncated AR protein product. The key domains shared among wild-type full-length AR (AR-FL) and all AR-Vs are the NH2-terminal transactivating domain (NTD) and DNA-binding domain (DBD). However, AR-Vs lack variable portions of the COOH-terminal domain including the ligand-binding domain (LBD).19,20,21 In spite of the fact that AR-Vs are unable to bind a ligand, they are constitutively active as transcription factors and capable of activating target genes.22
To date, at least 22 AR-Vs have been discovered in CRPC specimens.23 AR-V7 is the most clinically relevant variant as it is most frequently observed and the most abundant AR-V in clinical specimens. In addition, AR-V7 is the only variant that can be detected reproducibly at both the mRNA and protein levels. Moreover, detection of AR-V7 mRNA in circulating tumour cells (CTCs) and peripheral whole blood from CRPC patients treated with enzalutamide or abiraterone has been implicated in primary resistance and shorter progression-free and overall survival.24,25,26,27 Interestingly, the prevalence of AR-V7 was shown to be higher in enzalutamide-treated men who had previously received abiraterone and in abiraterone-treated men who had previously received enzalutamide.25 These findings were supported by an independent study that also utilised CTC-based RT-PCR assay.28 In this prospective study, it was shown that PSA response rate to abiraterone or enzalutamide was 7% among AR-V7-positive patients and 63% among AR-V7-negative patients. Another recent study demonstrated that AR-V7 detection in plasma-derived exosomal RNA strongly predicts resistance to enzalutamide or abiraterone in CRPC patients.29 Although these studies implicate that AR-V7 could be used as a treatment-specific biomarker, it is likely that other AR-Vs also play a role in the development of CRPC. For example, it was recently reported that AR-V9 is often co-expressed with AR-V7 in CRPC metastases, and predicts primary resistance to abiraterone.30
Recently, genomic structural rearrangements of AR (AR-GSRs) were established as a new class of AR gene alteration occurring in one third of CRPC-stage specimens.31 This work showed that the presence of AR-GSRs at high variant allele frequency was associated with outlier, tumour-specific expression of rearrangement-dependent AR-V species that displayed androgen-independent and enzalutamide-resistant transcriptional activity. However, contrary to the prior studies in cell lines,32,33 AR-GSRs were not associated with the AR-V7 expression levels in metastatic CRPC tissue.31 Another recent study utilising peripheral blood collected from patients with CRPC detected intra-AR structural variation in 15/30 patients of whom 14 expressed AR-Vs.34 Of note, most of the AR-V -positive patients expressed multiple AR-Vs, with AR-V7 being the most frequently occurring splice variant. However, AR-V3 was the most abundantly expressed AR splice variant. According to this study the presence of any AR-V was associated with shorter progression-free survival after second-line endocrine treatment compared to patients that did not harbour AR-Vs.34 Furthermore, in another recent investigation AR-GSRs in circulating tumour DNA were shown to associate with primary resistance to enzalutamide or abiraterone also in treatment-naïve CRPC patients with metastatic disease.13
Our aim was to study AR splice variants, rearrangements, mutations and copy-number variations (CNVs) in different stages of PC to better understand the emergence of CRPC. We used multiple sample cohorts representing hormone-naïve PCs and lymph node metastases as well as locally recurrent and metastatic CRPCs. We first employed whole-genome and whole-transcriptome sequencing followed by targeted AR sequencing panels allowing deeper sequence coverage. In particular, our aim was to confirm whether AR-Vs are expressed in higher levels in CRPC samples compared to earlier stage cancers. In addition, we wished to elucidate to what extent AR-V expression is due to the aberrant splicing and, on the other hand, AR gene rearrangements. We also wanted to study the association between AR-V and AR-FL expression; and to find out whether AR-V expression affects the expression of AR-regulated genes.
Materials and methods
Sample sets
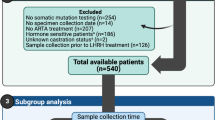
Two different sample sets utilised in the study are shown in Table 1 and are described in more detail in Supplementary Table S1. The sample set 1 contained freshly frozen tissue specimens from benign prostatic hyperplasia (BPH) (n = 12), hormone-naïve PC (n = 30) and locally recurrent CRPC (n = 13) with clinicopathological characteristics of PC cases and prior treatments of CRPC cases being shown in Supplementary Table S1. BPH samples were obtained by radical prostatectomy, cystoprostatectomy and by transurethral resection of the prostate. Hormone-naïve PC samples were obtained by radical prostatectomy and locally recurrent CRPCs by transurethral resection of the prostate. Histological evaluation and Gleason grading were performed by a pathologist based on haematoxylin/eosin stained slides. All samples contained a minimum of 70% cancerous or hyperplastic cells. DNA and RNA were isolated simultaneously using an AllPrep DNA/RNA Mini Kit (Qiagen, Valencia, CA, USA), according to manufacturer’s protocol. For certain samples, additional total RNA was isolated using Trizol (Invitrogen, Carlsbad, CA, USA) extraction according to manufacturer’s protocol. Three CRPC samples had RNA extracted using both Trizol and Qiagen AllPrep. Integrity was checked using Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
The sample set 2 consisted of 24 additional hormone-naïve PCs removed by prostatectomy, of which six specimens were also included in sample set 1, eight lymph node metastases obtained at lymphadenectomy and 30 metastatic CRPC specimens obtained at autopsy (clinicopathological characteristics of the cases are shown in Supplementary Table S1). Hormone-naïve PC samples contained a minimum of 60% cancerous or hyperplastic cells and were processed as described in the previous section.
Portions of the metastatic cancer tissue from pelvic lymphatic metastasis obtained at lymphadenectomy were used for this study. None of the eight patients had undergone ADT, chemotherapy or radiation therapy prior to this surgery. Precise histological control was achieved for all tissues studied in this group using the following protocol. Serial cryostat sectioning was used to identify portions of the sample containing a lower fraction of tumour cells. These areas were manually microdissected from the tissue block every 300 µm based on H&E stained slide visual analysis. The tumour cell fraction was 70% or greater by histologic visual estimation. DNA purification was performed as described previously.35 Total RNA was isolated using an AllPrep DNA/RNA/miRNA Universal Kit (Qiagen, Valencia, CA, USA) according to manufacturer’s instructions. The integrity of isolated RNA was confirmed using Fragment Analyzer (Advanced Analytical Technologies, Ankeny, IA, USA).
Metastatic CRPC specimens were obtained from 30 men who participated in the PELICAN (Project to ELIminate lethal CANcer) integrated clinical-molecular autopsy study of metastatic PC (a detailed sample list is shown in Supplementary Table S1). Androgen axis and corticosteroid clinical treatments are listed in the Supplementary Table S1. All metastases (one metastasis per patient) and noncancerous (normal, NL) control samples studied were obtained at autopsy. Isolated frozen tissue samples were serial cryostat microdissected for histological tumour purity >75%, and high-molecular-weight DNA was isolated using proteinase K digestion and phenol/chloroform extraction. Total RNA was isolated using an AllPrep DNA/RNA/miRNA Universal Kit (Qiagen, Valencia, CA, USA) according to manufacturer’s instructions. The integrity of isolated RNA was confirmed using Fragment Analyzer (Advanced Analytical Technologies, Ankeny, IA, USA).
Low-coverage (4–6×) whole-genome DNA sequencing and whole-transcriptome sequencing (applied to sample set 1) have been described before.36
Targeted AR DNA assay library construction and sequencing (applied to sample set 2)
A custom DNA sequencing panel was designed to cover all AR exons and introns. In addition, FOXA1 exons and SPOP exons 6-7 were included in the panel. Targeted sequence enrichment was performed using the SureSelectXT Target Enrichment System (Agilent Technologies, Santa Clara, CA, USA) according to manufacturer’s instructions. Briefly, 200 ng of genomic DNA was fragmented using Covaris® (Covaris, MA, USA) to yield a fragment size of 150–200 bp. End repair, addition of the 3’-dA overhang, ligation of indexing-specific adaptors, hybridisation to custom RNA baits, hybrid capture selection and index tagging were performed according to the Illumina paired-end sequencing library protocol. All recommended quality controls were performed between steps. The multiplexed samples were sequenced on the Illumina Miseq platform using 150 bp paired-end reads.
Targeted AR RNA assay library construction and sequencing (applied to sample set 2)
AR and five androgen-responsive genes (KLK3, FKBP5, TMPRSS2, ACPP and SLC45A3) were targeted for capture and sequencing. In addition, three house-keeping genes TBP, STARD7 and DDX1 were included for normalisation purposes. This custom RNA sequencing panel was designed to cover all AR exons and nonrepetitive intronic regions to enable investigation of most common AR splicing variants (AR-V3, AR-V4, AR-V5, AR-V6, AR-V7, AR-V9, AR-V12 and AR-45); other genes were covered less intensively (one or five amplicons per gene). Targeted sequence enrichment was performed using the SureSelectXT RNA Target Enrichment System (Agilent Technologies, Santa Clara, CA, USA) according to manufacturer’s instructions. Briefly, poly(A) RNA was purified from 1 µg of total RNA and fragmented chemically. In the following steps, samples were prepared using SureSelect Strand-Specific RNA Library Prep Kit to obtain adaptor-ligated cDNA library amplicons. Finally, hybridisation to custom RNA baits, hybrid capture selection and index tagging were performed. All the AMPure XP bead purification steps were conducted as instructed. The multiplexed samples were sequenced on the Illumina Miseq platform using 150 bp paired-end reads. The following modifications were made to the protocol if RNA was highly degraded (RQN < 6 determined by Fragment Analyzer) as recommended by Agilent Technologies: (1) Instead of poly(A) RNA purification from 1 µg total RNA, Ribo-Zero Gold Magnetic Kit (Illumina, San Diego, CA, USA) was used to remove rRNA from 2 µg of total RNA. (2) Instead of fragmenting the purified RNA at 94 °C for 8 min, RNA was denatured at 65 °C for 5 min. (3) All AMPure XP bead purification steps were performed using 1.8:1 bead volume to sample volume ratio. (4) Instead of 13 cycles in the pre-capture PCR, the number of cycles was increased to 14.
Validation of the targeted sequencing panels
Targeted custom SureSelect sequencing panels were validated by evaluating their performance in detecting AR aberrations in comparison to our previously published whole-genome DNA-seq data37,38 and whole-transcriptome RNA-seq data from this study. There was a good concordance in mutation detection between SureSelect DNA panel and previously analysed data from 22Rv1 cell line sample and metastatic CRPC samples from patients A21, A22 and A24. SureSelect DNA-seq detected the previously found H875Y mutation from 22Rv1 cell line, L702H mutation from liver metastasis from patient A21 as well as T878A mutation from a pelvic lymph node metastasis from patient A22, and from a right rib metastasis from patient A24.37,38 The data from AR splicing variant analysis also showed good accordance between SureSelect RNA panel and whole-transcriptome RNA-seq in three PC cell lines and two patient samples (Supplementary Fig. S1). It should be noted that SureSelect RNA assay was more sensitive in detecting AR-V9 than whole-transcriptome RNA-seq.
Bioinformatics
For analysis of targeted DNA-seq data, Illumina MiSeq reads were aligned to GRCh37 (hg19) genome using Bowtie2.39 AR, FOXA1 and SPOP variants were called using an in-house pipeline that utilises samtools mpileup.40 Filtered variants were annotated using the ANNOVAR software.41 Variants in dataset 1 were analysed from the whole-transcriptome sequencing data similarly.
AR copy numbers were analysed by calculating aligned read counts within overlapping 400 bp windows along the targeted regions using bedtools.42 The median of all AR bait coverage ratios that were obtained by dividing each normalised bait coverage value by the median of all values was used as the estimate of AR copy number. Chromosomal rearrangements were called using the in-house Breakfast algorithm that looks for paired-end reads and individual mates overlapping a chromosomal breakpoint.
For AR splice variant analysis using targeted or WTS RNA-seq data, Illumina MiSeq reads or HiSeq reads were aligned to an indexed reference fasta file containing unique signature sequences for various AR-Vs and AR-FL. The signatures consisted of 130 bp of the 3’ end of upstream exon and 130 bp of the 5’ end of downstream exon of a given unique splice junction (Supplementary Table S2). Relative AR-V expression was estimated as the percentage of all AR transcripts by dividing the number of reads aligned to a given AR-V signature by the total number of reads aligning to all the splice junctions containing the same upstream exon.
Expression levels of known AR-regulated genes were determined by aligning RNA-seq reads to GRCh37 genome using TopHat2.43 Z-scores were calculated from the normalised read counts, and AR-signalling score was computed as the sum of the Z-scores of all AR-regulated genes. Full bioinformatics methods are described in the Supplementary methods.
Immunohistochemistry
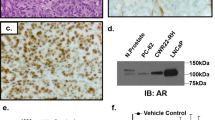
Formalin-fixed, paraffin-embedded tumour microarrays of hormone-naïve PC, locally recurrent CRPC and metastatic CRPC (described in Leinonen et al. 2013) were used. Immunohistochemistry for AR (with N-terminal antibody recognising full-length AR and the variants) has been previously described.44 For AR-V7-specific staining, sections were deparaffinised, and antigen retrieval was performed by using Tris-EDTA buffer 0.05% Tween-20 (pH 9) at +98 °C for 15 min. The staining was performed by Lab Vision Autostainer (ThermoFischer Scientific Inc., Waltham, MA, USA). The primary antibody Anti-Androgen Receptor (AR-V7 specific) Rabbit Monoclonal Antibody [RM7] (RevMAb Biosciences, San Francisco, CA, USA) and secondary antibody (N-Histofine® Simple Stain MAX PO; Nichirei, Tokyo, Japan) were used. ImmPACT DAB (Vector Laboratories, Burlingame, CA, USA) was used as a chromogen. The sections were counterstained with haematoxylin and mounted with DPX mounting medium (Sigma-Aldrich). The percentage of AR-V7-positive cells between PC and CRPC groups was statistically assessed with Mann-Whitney test.
Results
AR mutations and CNVs are detected only in CRPC cases
Since the mechanisms leading to emergence of CRPC are still largely unknown, we wanted to study the expression of AR splicing variants and other AR aberrations in tandem during different stages of PC. For this purpose, we performed low-coverage whole-genome DNA sequencing and whole-transcriptome sequencing in sample set 1 that included BPH specimens, hormone-naïve PC from prostatectomies and locally recurrent CRPCs (Supplementary Fig. S2). In addition, we performed targeted AR DNA and RNA sequencing in sample set 2 that contained hormone-naïve PC from prostatectomies and lymph node metastases as well as CRPC metastases (Fig. 1).
Combined DNA and RNA sequencing data from sample set 2 assayed by targeted SureSelect AR sequencing. AR mutations, copy-number alterations, summed score of AR-regulated gene expression and AR-V expression level as a fraction of AR transcript are shown. AR-V fractions are shown as CI95 lower bound values. Additionally, FOXA1 and SPOP mutation status is included
First, we wanted to analyse the status of AR mutations and CNVs across widely diverse set of samples to better understand their potential link to AR-V expression. As expected, AR mutations and CNVs were detected only in locally recurrent and metastatic CRPC specimens (Fig. 1 and Supplementary Fig. S2, upper panels). T878A mutation that has been shown to confer agonist activity of flutamide on the AR45,46,47 was found in 1/13 (8%) of locally recurrent CRPC specimens and in 2/23 (9%) of metastatic CRPC specimens. L702H mutation that converts glucocorticoids to AR agonists48,49 was found in 3/23 (13%) of metastatic CRPC specimens. Indeed, all three patients harbouring L702H mutation had been treated with glucocorticoids (Supplementary Table S1; a detailed treatment history is shown for patients having AR mutations). Copy-number gains (>1 copy of AR) or amplifications (>2 AR copies) were observed in 4/9 (44%) of locally recurrent CRPC specimens and in 19/23 (83%) of metastatic CRPC specimens, respectively (Fig. 1 and Supplementary Fig. S2). It should be noted that there were striking differences in AR copy numbers in metastatic CRPC specimens; for example, the lesion from patient A7 had six AR copies, whereas the lesion from patient A4 had as many as 68 AR copies. In four metastatic CRPC specimens, AR gain or amplification co-occurred with AR mutation. Since a large body of data, including our current investigation, has established that there are no mutations or copy number aberrations of AR in untreated PCs, majority of prostatectomy specimens in sample set 2 were not assayed with the targeted SureSelect DNA panel (Fig. 1). Additionally, data from targeted DNA assay are missing from those metastatic CRPC specimens of which DNA was not available (Fig. 1). The overall average coverage of the targeted regions in the samples ranged from 109× to 1829×, with the average coverage in the AR region being somewhat higher (114×–3358×).
The expression of AR-Vs is highest in CRPCs and associates with expression of AR-FL
Next we studied the presence of known AR-Vs that were detected from the RNA-seq data by aligning the reads against indexed AR-V signature sequence file containing exon-exon junction sequences unique to every AR-V under investigation (an example of RNA-seq read alignment of patient A17 is visualised in Supplementary Fig. S3). The AR-Vs detected by our assays included AR-V3, AR-V4, AR-V5, AR-V6, AR-V7 and AR-V9. The expression levels of AR-V4, AR-V5 and AR-V6 were negligible in comparison to AR-V3, AR-V7 and AR-V9, and were mainly observed in CRPC metastases. In sample set 1 run by whole-transcriptome RNA-seq, BPH specimens were mainly devoid of AR-V expression. Instead, AR-V3 and AR-V7 expression were detected in both hormone-naïve PC from prostatectomies and locally recurrent CRPCs with minimal co-expression of AR-V9 (Supplementary Fig. S2). Whereas the expression of AR-V3 was quite similar in the two different categories of samples, higher AR-V7 expression levels were detected in locally recurrent CRPCs as compared with hormone-naïve PC from prostatectomies. Since the depth of the whole-transcriptome sequencing was not satisfactory (average per-base sequence coverage ranged from 14×–137×) in terms of reliable detection of AR variants, we also performed targeted RNA sequencing of the AR, which provided an average coverage range from 95× to 2247× utilising sample set 2 (Fig. 1). In sample set 2, not only the expression level of AR-V7 but also the expression levels of AR-V3 and AR-V9 were higher in metastatic lesions from CRPC cases compared to hormone-naïve PC from prostatectomy (Fig. 1). The differences were statistically significant for either variant alone (Supplementary Table S3) or when their expression fractions were combined (p = 0.0006, unpaired Wilcoxon rank sum test, Table 2). In addition, metastatic CRPC cases expressed significantly more AR-V3, AR-V7 and AR-V9 compared to non-androgen-deprived pelvic lymph node metastases (p = 0.0282, unpaired Wilcoxon rank sum test, Table 2). We also studied whether the expression of AR-Vs is associated with the CNV status (neutral vs. duplicated/amplified AR) in sample set 2. There was a modest correlation when CNV status was compared to the combined expression levels of AR-V3, AR-V7 and AR-V9 (rho = 0.39, p = 0.005, Spearman’s rank correlation).
In sample set 2, AR-FL expression was threefold higher in metastatic CRPC compared to hormone-naïve PC from prostatectomy specimens, whereas in sample set 1, AR-FL was expressed fivefold higher in CRPC lesions than in prostatectomy samples. Notably, the expression of AR-V3, AR-V7 and AR-V9 was strongly associated with the levels of full-length AR in sample set 2 (Fig. 2) and in sample set 1 (Supplementary Fig. S4), suggesting that the expression of AR locus drives the expression of AR-Vs both in hormone-naïve PC and in CRPC. Furthermore, there was strong and highly significant correlation between the expression of each individual AR-V compared to other AR-Vs in sample set 2 (Fig. 3).
The correlation between AR-FL mRNA expression and mRNA expression of (a) AR-V3, (b) AR-V7, (c) AR-V9, (d) all three AR-Vs combined utilising specimens from sample set 2. The counts of splice junction reads indicative of AR-FL or AR-Vs are plotted in the y-axis and x-axis, respectively. Spearman’s rank correlation coefficients and p values computed via the asymptotic t approximation are also shown in the figures
The correlation between (a) AR-V7 and AR-V9 mRNA expression, (b) AR-V7 and AR-V3 mRNA expression and (c) AR-V9 and AR-V3 mRNA expression utilising specimens from sample set 2. The counts of splice junction reads indicative of given AR-Vs are plotted in the y-axis and x-axis. Spearman’s rank correlation coefficients and p values computed via the asymptotic t approximation are also shown in the figures
We also asked whether AR variant expression affects the expression of AR-regulated genes. This was done by calculating the summed z-score of five androgen-responsive genes (KLK3, FKBP5, TMPRSS2, ACPP and SLC45A3) (Fig. 1, Supplementary Fig. S2). AR-V expression was not associated with AR-regulated gene expression in sample set 2 when the proportion of fractions, when compared to AR-FL, of each AR-V or all AR-Vs combined, were plotted against AR signalling score (Supplementary Fig. S5). In addition, we wanted to test whether AR-V3, AR-V7 or AR-V9 expression correlates in particular to KLK3 expression. No correlation between either variant and KLK3 was detected in metastatic CRPC specimens (Supplementary Fig. S6). We next studied mutation status of two AR-regulating genes, FOXA1 and SPOP, in sample set 2. FOXA1 mutations were found in 3/8 (38%) lymph node metastases and in 1/23 (4%) CRPC metastases, whereas SPOP mutations were detected in 1/8 (13%) lymph node metastases and in 2/23 (9%) CRPC metastases (Fig. 1). We did not find any association between FOXA1 or SPOP mutation status and AR-regulated gene expression. All mutations found in this study and their variant allele frequencies are shown in Supplementary Table S4.
AR genomic structural rearrangements occur in the context of amplified AR
AR genomic structural rearrangements (AR-GSRs) were recently identified as a novel class of AR alteration using both autopsy CRPC specimens and peripheral blood collected from CRPC patients.31,34 More importantly, the presence of AR-GSRs was associated with expression of AR-Vs in both studies. To this end, we analysed AR DNA-seq data with our structural variant detection pipeline to identify AR-GSRs, defined as events having at least one breakpoint detected within the AR gene region. Average per-base sequence coverage of the AR gene region ranged from 114×–3358× and on average 78% of AR was covered by at least 10 reads (range 76–82%). We detected putative AR-GSRs in 5/30 metastatic CRPC patients who all harboured a highly amplified AR (Supplementary Table S5). All other sample types were negative for AR-GSRs when cut-off of 10 supporting split reads was used. It should be noted that none of the AR-GSRs occurred along with AR missense mutations. The break fusion junctions of AR-GSRs were variable, demonstrating several types of rearrangements including duplication, deletion, inversion and translocation events. Furthermore, all patients demonstrated unique AR-GSR breakpoint locations. Interestingly, patient A27 displayed a rearrangement that deleted half of exon 4 as well as exons 5 and 6 and was the only patient whose AR-GSR was also detected from the RNA-seq sample (Supplementary Fig. S7). This rearrangement may lead to translation of a truncated, constitutively active protein product, and could thus have some biological relevance. None of the AR-GSRs detected by our pipeline were associated with the expression of previously known AR-Vs and their variant allele fractions were relatively low (range 2.6–10.9%).
Expression of AR-V7 is heterogeneous at the protein level
To study how the detected differences in AR variant expression between PC stages are translated to the protein level, we performed immunohistochemistry against AR-V7 with tumour microarrays of hormone-naïve PC from prostatectomies (n = 146), locally recurrent CRPCs (n = 97) and metastatic CRPC samples (103 metastases in total from 31 patients; 1–5 metastases per patient). We also studied immunohistochemistry of AR (N-terminal antibody recognising full-length AR as well as all variants containing exon 1, including AR-V3, AR-V7 and AR-V9). As a positive control we used a sample of 22Rv1 cell line known to contain high AR-V7 expression (Supplementary Fig. S8a). Primarily, AR-V7 was detected in the nucleus (92% of hormone-naïve, 62% of CRPC and 75% of metastatic samples), although variable cytoplasmic staining could be detected in minority of the samples in all phases of the disease (12% of hormone-naïve, 32% of CRPC and 21% of metastatic samples) (Supplementary Fig. S8b). In contrast to AR staining, the AR-V7 staining was heterogeneous and often present in only a fraction of the cells (Supplementary Fig. S8b). For example, 89% of the positive, hormone-naïve cases had nuclear AR-V7 in less than 10% of the cells (mean value of positive cells 6.4%, median 3.2%) (Supplementary Fig. S9a,b). In CRPC, the number of AR-V7-negative cases increased as compared to hormone-naïve disease (38% vs. 8% of no nuclear AR-V7 detected, respectively) (Supplementary Fig. S9a). Interestingly, many of the AR-V7-negative CRPC samples had a strong mesenchymal phenotype, while the cells in most positive tumours had round, epithelial phenotype. In the positive CRPC cases, the percentage of AR-V7-positive cells increased as compared to hormone-naïve disease, with mean value 24.9% and median 13.6% (Supplementary Fig. S9b). As for the metastatic disease, 88% of the tumours studied had detectable AR-V7 positivity, and all 31 patients had one or more AR-V7-positive metastases. It should be noted that direct comparison of AR-V7 mRNA and protein levels is not possible in most of the cases as samples do not originate from the same tumour areas. However, general observations can be made. For example, patient A28 with highest AR-V7 expression at the mRNA level in the metastasis subjected to sequencing analysis (Fig. 1) had AR-V7 positivity in all four metastases that were studied with immunohistochemistry.
Discussion
This study describes the AR aberration status in two comprehensive patient cohorts including specimens from BPH, untreated localised and metastatic PC as well as both locally recurrent and metastatic CRPCs. We show that even though AR-V3, AR-V7 and AR-V9 are expressed widely in different sample types, they are statistically more highly expressed in metastatic CRPCs in comparison to two hormone-naïve sample groups, prostatectomies and lymph node metastases. This further reinforces the conception that AR-Vs likely have a role in CRPC progression and development of resistance to AR-targeted therapies.
In CRPC metastases, the expression of AR-V7 was 13% of AR transcript at maximum, and it was present in 21/29 cases, whereas the expression levels of AR-V3 and AR-V9 were highly similar (7% of AR transcript at maximum) and detected in 23/29 and 22/29 CRPC metastases, respectively. Our finding that AR-V3, AR-V7 and AR-V9 are present at varying levels also in benign prostate tissue and hormone-naïve primary PCs is in line with previous reports.23,50 Furthermore, our whole-genome and targeted DNA sequencing results were in accordance with previous reports demonstrating that AR mutations and amplifications are rare in early stages of untreated PC, but occur much more frequently in patients affected by metastatic CRPC.4,23,50 In our study, no AR mutations or copy number changes were detected in untreated cases; they were observed only in locally recurrent and metastatic CRPC specimens. In CRPC metastases, 5/23 cases harboured an AR mutation and 19/23 cases had a copy number gain or amplification. Out of 22 cases of metastatic CRPC of which both DNA- and RNA-seq data were available, all but one patient (patient A5) had at least one AR aberration underlining the crucial role of AR in the disease progression. Figure 4 summarises both genome and RNA level alterations of AR detected in this study during different stages of PC.
Summary of the frequency of the genome and RNA level alterations of AR during different stages of prostate cancer. Copy number (CN) changes of AR are presented as gains (>1 copy of AR) and amplifications (>2 AR copies). AR-V expression levels are divided into AR-V low (<5% of splice variant of AR transcript) and AR-V high (>5% of splice variant of AR transcript) groups. The data are from MiSeq assays for all other sample groups, except for BPH and locally recurrent CRPC whose data are from HiSeq assays
It is noteworthy that lymph node metastasis specimens from patients who had not undergone any ADT did not show elevated levels of AR-Vs. It has been demonstrated earlier using several PC cell lines that inhibition of the full-length AR protein via castration, antiandrogen treatment or siRNA induced the expression of AR-V7, although concomitant, yet lesser increases in full-length AR were also observed.51,52 It has also been shown that ADT does not directly regulate levels of AR-V7, but rather enhances AR gene transcription rate and splicing factor recruitment to AR pre-mRNA, thus elevating AR-V7 levels.53 Accordingly, we also showed that the expression levels of AR-V7 as well as levels of AR-V3 and AR-V9 were strongly associated with the levels of full-length AR, indicating that the AR-V expression is dependent on transcription rate of AR locus.
Interestingly, we observed that AR-V7 was strongly co-expressed with AR-V9 in sample set 2, which is in line with a recent report also demonstrating simultaneous expression of AR-V7 and AR-V9 in CRPC metastases.30 Moreover, our data showed that AR-V7 was co-expressed with AR-V3; and there was also a clear positive correlation between expression of AR-V9 and AR-V3. It should be noted that out of 25 CRPC metastases that expressed any AR variant, as many as 17 cases harboured expression of all three of these AR-Vs. Since AR-V3, AR-V7 and AR-V9 are constitutively active, it is reasonable to expect that their combined contribution to PC progression might be greater than what could be expected when their effects are studied separately. In our data, AR-V expression levels were above 5% when compared to overall AR transcript expression levels in 11 metastatic CRPC specimens (sample set 2). Since metastatic CRPC specimens harboured three times higher expression of AR-FL in comparison to hormone-naïve prostatectomy samples it would mean that 5% AR-V fraction does not yet bring the AR-V levels to corresponding levels of AR-FL in hormone-naïve PC. However, the levels of AR-V required to drive an androgen-independent transcriptome are unknown.
It has been demonstrated in several cell line studies that AR-Vs are able to induce the expression of AR-controlled genes such as KLK3, TMPRSS2 and FKBP5 in the absence of androgens or AR-FL.19,20,54,55 Therefore, we interrogated the levels of classical AR-regulated genes in our sample sets, and calculated the summed z-score of five androgen responsive genes (KLK3, FKBP5, TMPRSS2, ACPP and SLC45A3). There was no association between the expression levels of AR-V3, AR-V7 or AR-V9 and z-score in sample set 2. Likewise, no correlation was detected when KLK3 expression was compared to the expression levels of these AR-Vs in CRPC metastases. One explanation for this discrepancy could be the fact that metastatic CRPC samples expressing the highest levels of AR-Vs were taken at autopsy making it highly unlikely that AR-regulation was anymore classical at the late stage of the disease. In addition, it has previously been demonstrated that bone metastases with high AR-V levels did not show high levels of KLK3, KLK2, FKBP5, TMPRSS2 and NKX3-1, whereas the levels of other transcripts known to be positively regulated by AR were elevated (including CDK1, CYCLINA2, HSP27 and C-MYC).56 Therefore, it seems that the expression profile induced by AR-Vs can be context-dependent and might not correspond to the effects observed in cell lines.
As protein expression does not always fully correlate with mRNA expression, and as the AR-Vs may also be regulated post-transcriptionally, it is of importance to study their expression also at the protein level. We assessed the expression of AR-V7 by immunohistochemistry in hormone-naïve PC, locally recurrent CRPC and metastatic CRPC samples. Although a third of CRPCs in this cohort were found negative for AR-V7 protein, the results support the view that AR-V7 expression increases during castration resistance, and that the protein is present in most PC metastases. It is noteworthy that expression of AR-V7 is highly more heterogeneous than that of AR overall. This indicates either differences in transcriptional expression of AR-V7 between tumour cells, or heterogeneous post-transcriptional regulation of it within tumour cell populations.
Genomic structural rearrangements (GSRs) have recently been shown to define a class of AR aberrations occurring at a considerable frequency in CRPC material.31,34 Henzler et al. studied AR-GSRs in 30 rapid autopsy CRPC soft tissue metastases obtained from 15 patients and found that 10/30 metastases (6/15 patients) displayed at least one AR-GSR event. Instead, De Laere et al. utilised liquid biopsies from 30 chemotherapy pretreated or chemo-naïve CRPC patients and detected at least one AR-GSR in 15/30 patients. We observed AR-GSRs in 5/30 patients with metastatic CRPC, which is considerably less when compared to these prior findings. The reason for this discrepancy is unclear, but it can be at least partly due to the fact that there were more uncovered regions in our assay (76–82% of AR covered) than in Henzler et al. assay (83–89% of AR covered). Interestingly, AR-GSRs were not detected in the context of AR missense mutations in both our and Henzler et al. sample cohorts.31 Furthermore, in our material, the break fusion junctions of AR-GSRs were variable, demonstrating several types of rearrangements, but none of the AR-GSRs were associated with the expression of previously known AR-Vs. Patient A27 was the only one whose AR-GSR was also detected by RNA-seq and his variant allele fraction was also the highest being 10.9%. For other patients with AR-GSRs the variant allele fractions ranged from 2.6 to 8.6%. In CRPC metastases, half of the AR-GSR-positive patients expressed AR-Vs,31 whereas all but one of AR-GSR-positive patients who were liquid-biopsied harboured AR-V expression.34 Together, these results demonstrate that the connection of AR-GSRs and the expression of AR-Vs is highly variable in different sample cohorts. It is also noteworthy that in the study of Henzler et al., the only previously reported variant that was associated with the presence of AR-GSRs were AR-V7 and AR-V12 (ARv567es), but the data from De Laere et al. showed that the majority of AR-GSR-positive patients expressed multiple previously reported AR-Vs. AR-GSRs were restricted to CRPC specimens in both our and Henzler et al. data, suggesting that they are yet another means of CRPC to retain AR signalling.
In conclusion, the finding that AR-V expression levels increase in patients treated with androgen-deprivation therapy might indicate that there is a clonal selection pressure on the different tumour clones in order to maintain functional AR signalling independent of the androgen levels. We provide evidence that AR-V3, AR-V7 and AR-V9 are co-expressed in metastatic CRPC highlighting the fact that targeting of the AR ligand-binding domain might not be sufficient to achieve clinically relevant treatment responses. Consequently, inhibiting AR function via regions common to all AR-Vs is likely to provide additional benefit to patients with CRPC.
References
Visakorpi, T. et al. In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat. Genet. 9, 401–406 (1995).
Waltering, K. K. et al. Increased expression of androgen receptor sensitizes prostate cancer cells to low levels of androgens. Cancer Res. 69, 8141–8149 (2009).
Brooke, G. N., & Bevan, C. L. The role of androgen receptor mutations in prostate cancer progression. Curr. Genom. 10, 18–25 (2009).
Grasso, C. S. et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 487, 239–243 (2012).
Cai, C. & Balk, S. P. Intratumoral androgen biosynthesis in prostate cancer pathogenesis and response to therapy. Endocr. Relat. Cancer 18, R175–R182 (2011).
Locke, J. A. et al. Androgen levels increase by intratumoral de novo steroidogenesis during progression of castration-resistant prostate cancer. Cancer Res. 68, 6407–6415 (2008).
Montgomery, R. B. et al. Maintenance of intratumoral androgens in metastatic prostate cancer: a mechanism for castration-resistant tumor growth. Cancer Res. 68, 4447–4454 (2008).
Palmberg, C. et al. Androgen receptor gene amplification at primary progression predicts response to combined androgen blockade as second line therapy for advanced prostate cancer. J. Urol. 164, 1992–1995 (2000).
Gottlieb, B., Beitel, L. K., Nadarajah, A., Paliouras, M., & Trifiro, M. The androgen receptor gene mutations database: 2012 update. Hum. Mutat. 33, 887–894 (2012).
Eisermann, K., Wang, D., Jing, Y., Pascal, L. E., & Wang, Z. Androgen receptor gene mutation, rearrangement, polymorphism. Transl. Androl. Urol. 2, 137–147 (2013).
Attard, G. et al. Selective inhibition of CYP17 with abiraterone acetate is highly active in the treatment of castration-resistant prostate cancer. J. Clin. Oncol. 27, 3742–3748 (2009).
Tran, C. et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science 324, 787–790 (2009).
Annala, M. et al. Circulating tumor DNA genomics correlate with resistance to abiraterone and enzalutamide in prostate cancer. Cancer Discov. 8, 444–457 (2018).
Romanel, A. et al. Plasma AR and abiraterone-resistant prostate cancer. Sci. Transl. Med 7, 312re10 (2015).
Azad, A. A. et al. Androgen receptor gene aberrations in circulating cell-free DNA: biomarkers of therapeutic resistance in castration-resistant prostate cancer. Clin. Cancer Res. 21, 2315–2324 (2015).
Wyatt, A. W. et al. Genomic alterations in cell-free DNA and enzalutamide resistance in castration-resistant prostate cancer. JAMA Oncol. 2, 1598–1606 (2016).
Conteduca, V. et al. Androgen receptor gene status in plasma DNA associates with worse outcome on enzalutamide or abiraterone for castration-resistant prostate cancer: a multi-institution correlative biomarker study. Ann. Oncol. 28, 1508–1516 (2017).
Scher, H. I. et al. Antitumour activity of MDV3100 in castration-resistant prostate cancer: a phase 1-2 study. Lancet 375, 1437–1446 (2010).
Dehm, S. M., Schmidt, L. J., Heemers, H. V., Vessella, R. L., & Tindall, D. J. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 68, 5469–5477 (2008).
Hu, R. et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 69, 16–22 (2009).
Lu, C., & Luo, J. Decoding the androgen receptor splice variants. Transl. Androl. Urol. 2, 178–186 (2013).
Jenster, G. et al. Domains of the human androgen receptor involved in steroid binding, transcriptional activation, and subcellular localization. Mol. Endocrinol. 5, 1396–1404 (1991).
Robinson, D. et al. Integrative clinical genomics of advanced prostate cancer. Cell 161, 1215–1228 (2015).
Antonarakis, E. S. et al. Androgen receptor splice variant 7 and efficacy of taxane chemotherapy in patients with metastatic castration-resistant prostate cancer. JAMA Oncol. 1, 582–591 (2015).
Antonarakis, E. S. et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N. Engl. J. Med. 371, 1028–1038 (2014).
Antonarakis, E. S. et al. Clinical significance of androgen receptor splice variant-7 mRNA detection in circulating tumor cells of men with metastatic castration-resistant prostate cancer treated with first- and second-line abiraterone and enzalutamide. J. Clin. Oncol. 35, 2149–2156 (2017).
Seitz, A. K. et al. AR-V7 in peripheral whole blood of patients with castration-resistant prostate cancer: association with treatment-specific outcome under abiraterone and enzalutamide. Eur. Urol. 72, 828–834 (2017).
Steinestel J., et al. Detecting predictive androgen receptor modifications in circulating prostate cancer cells. Oncotarget 2015. https://doi.org/10.18632/oncotarget.3925.
Del Re M, et al. The detection of androgen receptor splice variant 7 in plasma-derived exosomal RNA strongly predicts resistance to hormonal therapy in metastatic prostate cancer patients. Eur. Urol. 71, 680–687 (2017).
Kohli M., et al. Androgen receptor variant AR-V9 is coexpressed with AR-V7 in prostate cancer metastases and predicts abiraterone resistance. Clin Cancer Res. 23, 4704–4715 (2017).
Henzler, C. et al. Truncation and constitutive activation of the androgen receptor by diverse genomic rearrangements in prostate cancer. Nat. Commun. 7, 13668 (2016).
Li, Y. et al. Intragenic rearrangement and altered RNA splicing of the androgen receptor in a cell-based model of prostate cancer progression. Cancer Res. 71, 2108–2117 (2011).
Li, Y. et al. AR intragenic deletions linked to androgen receptor splice variant expression and activity in models of prostate cancer progression. Oncogene 31, 4759–4767 (2012).
De Laere B., et al. Comprehensive profiling of the androgen receptor in liquid biopsies from castration-resistant prostate cancer reveals novel intra-AR structuralvariation and splice variant expression patterns. Eur. Urol. 72, 192–200 (2017).
Bova, G. S. et al. Homozygous deletion and frequent allelic loss of chromosome 8p22 loci in human prostate cancer. Cancer Res. 53, 3869–3873 (1993).
Annala, M. et al. Recurrent SKIL-activating rearrangements in ETS-negative prostate cancer. Oncotarget 6, 6235–6250 (2015).
Gundem, G. et al. The evolutionary history of lethal metastatic prostate cancer. Nature 520, 353–357 (2015).
Bova, G. S. et al. Integrated clinical, whole-genome, and transcriptome analysis of multisampled lethal metastatic prostate cancer. Cold Spring Harb. Mol. Case Stud. 2, a000752 (2016).
Langmead, B., & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Quinlan, A. R., & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Leinonen, K. A. et al. Loss of PTEN is associated with aggressive behavior in ERG-positive prostate cancer. Cancer Epidemiol. Biomark. Prev. 22, 2333–2344 (2013).
Lallous, N. et al. Functional analysis of androgen receptor mutations that confer anti-androgen resistance identified in circulating cell-free DNA from prostate cancer patients. Genome Biol. 17, 10 (2016).
Zhou, J., Liu, B., Geng, G., & Wu, J. H. Study of the impact of the T877A mutation on ligand-induced helix-12 positioning of the androgen receptor resulted in design and synthesis of novel antiandrogens. Proteins 78, 623–637 (2010).
Taplin, M. E. et al. Selection for androgen receptor mutations in prostate cancers treated with androgen antagonist. Cancer Res. 59, 2511–2515 (1999).
Carreira, S. et al. Tumor clone dynamics in lethal prostate cancer. Sci. Transl. Med 6, 254ra125 (2014).
Zhao, X. Y. et al. Glucocorticoids can promote androgen-independent growth of prostate cancer cells through a mutated androgen receptor. Nat. Med. 6, 703–706 (2000).
Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate. Cancer Cell. 163, 1011–1025 (2015).
Yu, Z. et al. Rapid induction of androgen receptor splice variants by androgen deprivation in prostate cancer. Clin. Cancer Res. 20, 1590–1600 (2014).
Hu, R. et al. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res. 72, 3457–3462 (2012).
Liu, L. L. et al. Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene 33, 3140–3150 (2014).
Guo, Z. et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 69, 2305–2313 (2009).
Sun, S. et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J. Clin. Invest. 120, 2715–2730 (2010).
Hornberg, E. et al. Expression of androgen receptor splice variants in prostate cancer bone metastases is associated with castration-resistance and short survival. PLoS One 6, e19059 (2011).
Acknowledgements
We are grateful to Orion for providing the AR-V7 specific antibody. We thank Päivi Martikainen, Riina Kylätie and Marika Vähä-Jaakkola for their technical assistance. The work was supported in parts by grants from the Finnish Funding Agency for Technology and Innovation, the Academy of Finland, the Cancer Society of Finland, the Sigrid Juselius Foundation and the Medical Research Fund of Tampere University Hospital.
Author contributions
H.M.L.K. wrote the article and conducted the NGS laboratory experiments. R.H. and M.A. conducted the computational analyses. L.L. performed the IHC stainings and analyses. A.B. helped with the NGS laboratory experiments. K.K., M.N., H.G.L. and T.V. designed the study. T.L.T., W.B.I. and G.S.B. provided materials. H.M.L.K. and T.V. analysed the data. All authors read and approved the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The use of benign prostatic hyperplasia, hormone-naïve prostate cancer and locally recurrent CRPC specimens was approved by the ethical committee of the Tampere University Hospital. Written informed consent was obtained from the subjects. Samples from non-androgen deprived pelvic lymph node metastases were obtained under Johns Hopkins Medicine IRB approval 04-05-03-02. Metastatic CRPC specimens were obtained from 30 men who participated in the PELICAN (Project to ELIminate lethal CANcer) integrated clinical-molecular autopsy study of metastatic prostate cancer. Subjects consented to participate in the Johns Hopkins Medicine IRB-approved study between 1995 and 2005. The study was performed in accordance with the Declaration of Helsinki.
Availability of data and material
All data generated or analysed during this study are included in this published article [and its supplementary information files].
Note
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution 4.0 International (CC BY 4.0).
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Kallio, H.M.L., Hieta, R., Latonen, L. et al. Constitutively active androgen receptor splice variants AR-V3, AR-V7 and AR-V9 are co-expressed in castration-resistant prostate cancer metastases. Br J Cancer 119, 347–356 (2018). https://doi.org/10.1038/s41416-018-0172-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-018-0172-0
This article is cited by
-
Co-expression and clinical utility of AR-FL and AR splice variants AR-V3, AR-V7 and AR-V9 in prostate cancer
Biomarker Research (2023)
-
Long-range gene regulation in hormone-dependent cancer
Nature Reviews Cancer (2023)
-
The promising role of new molecular biomarkers in prostate cancer: from coding and non-coding genes to artificial intelligence approaches
Prostate Cancer and Prostatic Diseases (2022)
-
Androgen receptor splice variant 7 detected by immunohistochemical is an independent poor prognostic marker in men receiving adjuvant androgen-deprivation therapy after radical prostatectomy
Biomarker Research (2021)
-
Androgen receptor gain in circulating free DNA and splicing variant 7 in exosomes predict clinical outcome in CRPC patients treated with abiraterone and enzalutamide
Prostate Cancer and Prostatic Diseases (2021)