Abstract
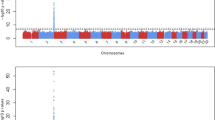
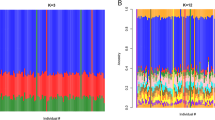
Studies in Caucasian and Asian populations consistently associated interindividual and interethnic variability in rosuvastatin pharmacokinetics to the polymorphisms SLCO1B1 c.521T>C (rs4149056 p. Val174Ala) and ABCG2 c.421C>A (rs2231142, p. Gln141Lys). To investigate the pharmacogenetics of rosuvastatin in African populations, we first screened 785 individuals from nine ethnic African populations for the SLCO1B1 c.521C and ABCG2 c.421CA variants. This was followed by sequencing whole exomes from individuals of African Bantu descent, who participated in a 20 mg rosuvastatin pharmacokinetic trial in Harare Zimbabwe. Frequencies of SLCO1B1 c.521C ranged from 0.0% (San) to 7.0% (Maasai), while ABCG2 c.421A ranged from 0.0% (Shona) to 5.0% (Kikuyu). Variants showing significant association with rosuvastatin exposure were identified in SLCO1B1, ABCC2, SLC10A2, ABCB11, AHR, HNF4A, RXRA and FOXA3, and appear to be African specific. Interindividual differences in the pharmacokinetics of rosuvastatin in this African cohort cannot be explained by the polymorphisms SLCO1B1 c.521T>C and ABCG2 c.421C>A, but appear driven by a different set of variants.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lee E, Ryan S, Birmingham B, Zalikowski J, March R, Ambrose H, et al. Rosuvastatin pharmacokinetics and pharmacogenetics in white and Asian subjects residing in the same environment. Clin Pharmacol Ther. 2005;78:330–41.
Pasanen MK, Neuvonen M, Neuvonen PJ, Niemi M. SLCO1B1 polymorphism markedly affects the pharmacokinetics of simvastatin acid. Pharm Genom. 2006;16:873–9.
SEARCH Collaborative Group. SLCO1B1 variants and statin-induced myopathy-a genome wide study. N Engl J Med. 2008;359:333–40.
Niemi M, Pasanen MK, Neuvonen PJ. Organic anion transporting polypeptide 1B1: a genetically polymorphic transporter of major importance for hepatic drug uptake. Pharmacol Rev. 2011;63:157–81.
DeGorter MK, Tirona RG, Schwarz UI, Choi Y-H, Dresser GK, Suskin N, et al. Clinical and pharmacogenetic predictors of circulating atorvastatin and rosuvastatin concentrations in routine clinical care. Circ Cardiovasc Genet. 2013;6:400–8.
Birmingham BK, Bujac SR, Elsby R, Azumaya CT, Zalikowski J, Chen Y, et al. Rosuvastatin pharmacokinetics and pharmacogenetics in Caucasian and Asian subjects residing in the United States. Eur J Clin Pharmacol. 2015;71:329–40.
1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74.
Chasman DI, Giulianini F, MacFadyen J, Barratt BJ, Nyberg F, Ridker PM. Genetic determinants of statin-induced low-density lipoprotein cholesterol reduction: The justification for the use of statins in prevention: An intervention trial evaluating rosuvastatin (JUPITER) trial. Circ Cardiovasc Genet. 2012;5:257–64.
Astra Zeneca. Crestor Prescribing Information. 2016. Accessed through https://www.accessdata.fda.gov/drugsatfda_docs/label/2010/021366s016lbl.pdf
Hoosain N, Pearce B, Jacobs C, Benjeddou M. Mapping SLCO1B1 genetic variation for global precision medicine in understudied regions in Africa: A focus on Zulu and Cape admixed populations. Omi A J Integr Biol. 2016;20:546–54.
Mpeta B, Kampira E, Castel S, Mpye KL, Soko ND, Wiesner L, et al. Differences in genetic variants in lopinavir disposition among HIV-infected Bantu Africans. Pharmacogenomics. 2016;17:679–90.
Soko N, Dandara C, Ramesar R, Kadzirange G, Masimirembwa C. Pharmacokinetics and pharmacogenetics of rosuvastatin in 30 healthy Zimbabwean individuals of African ancestry. Br J Clin Pharmacol. 2016;2:326–8.
Aklillu E, Habtewold A, Ngaimisi E, Yimer G, Mugusi S, Amogne W, et al. SLCO1B1 gene variations among Tanzanians, Ethopians and Europeans: relevance for African and worldwide precision medicine. OMICS. 2016;20:538–45.
Matimba A, Oluka MN, Ebeshi BU, Sayi J, Bolaji OO, Guantai AN, et al. Establishment of a biobank and pharmacogenetics database of African populations. Eur J Hum Genet. 2008;16:780–3.
Depristo MA, Banks E, Poplin RE, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. HHS Public Access. Nat Genet. 2011;43:491–8.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucl Acids Res. 2010;38:e164.
Thorn CF, Klein TE, Altman RB. PharmGKB: the pharmacogenomics knowledge base. Methods Mol Biol. 2013;1015:311–20.
Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucl Acids Res. 2016;44:D862–8.
Sifrim A, Popovic D, Tranchevent L-C, Ardeshirdavani A, Sakai R, Konings P, et al. eXtasy: variant prioritization by genomic data fusion. Nat Meth. 2013;10:1083–4.
Winter JCF. Using the Student’ s t-test with extremely small sample sizes. PARE. 2013;18:1–12.
Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–8.
Masimirembwa C, Hasler J. Pharmacogenetics in Africa, an opportunity for appropriate drug dosage regimens: on the road to personalized. Healthc CPT Pharmacomet Syst Pharmacol. 2013;2:e45.
Dandara C, Swart M, Mpeta B, Wonkam A, Masimirembwa C. Cytochrome P450 pharmacogenetics in African populations: implications for public health. Expert Opin Drug Metab Toxicol. 2014;10:769–85.
Soko ND, Masimirembwa C, Dandara C. Pharmacogenomics of Rosuvastatin: A glocal (global+local) African perspective and expert review on a statin drug. OMICS. 2016;20:498–509.
PCJL Santos, Soares RaG, Nascimento RM, Machado-Coelho GLL, Mill JG, Krieger JE, et al. SLCO1B1rs4149056 polymorphism associated with statin-induced myopathy is differently distributed according to ethnicity in the Brazilian general population: Amerindians as a high risk ethnic group. BMC Med Genet. 2011;12:136.
Ngaimisi E, Habtewold A, Minzi O, Makonnen E, Mugusi S, Amogne W, et al. Importance of ethnicity, CYP2B6 and ABCB1 genotype for Efavirenz pharmacokinetics and treatment outcomes: A parallel-group prospective cohort study in two sub-Saharan Africa populations. PLoS ONE. 2013;8:e67946.
Masimirembwa C, Dandara C, Leutscher PDC. Rolling out efavirenz for HIV precision medicine in Africa: are we ready for pharmacovigilance and tackling neuropsychiatric adverse effects? Omi A J Integr Biol. 2016;20:575–80.
Nebert DW. Extreme discordant phenotype methodology: an intuitive approach to clinical pharmacogenetics. Eur J Pharmacol. 2000;410:107–20.
Westlind-Johnsson A, Hermann R, Huennemeyer A, Hauns B, Lahu G, Nassr N, et al. Identification and characterization of CYP3A4*20, a novel rare CYP3A4 allele without functional activity. Clin Pharmacol Ther. 2006;79:339–49.
Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–25.
Apellaniz-Ruiz M, Lee M-Y, Sanchez-Barroso L, Gutierrez-Gutierrez G, Calvo I, Garcia-Estevez L, et al. Whole-exome sequencing reveals defective CYP3A4 variants predictive of paclitaxel dose-limiting neuropathy. Clin Cancer Res. 2015;21:322–8.
Zhang G, Nebert DW, Chakraborty R, Jin L. Statistical power of association using the extreme discordant phenotype design. Pharm Genom. 2006;16:401–13.
Martin PD, Warwick MJ, Dane AL, Hill SJ, Giles PB, Phillips PJ, et al. Metabolism, excretion, and pharmacokinetics of rosuvastatin in healthy adult male volunteers. Clin Ther. 2003;25:2822–35.
Cooper KJ, Martin PD, Dane AL, Warwick MJ, Raza A, Schneck DW. Lack of effect of ketoconazole on the pharmacokinetics of rosuvastatin in healthy subjects. Br J Clin Pharmacol. 2003;55:94–9.
Cooper KJ, Martin PD, Dane AL, Warwick MJ, Schneck DW, Cantarini MV. The effect of fluconazole on the pharmacokinetics of rosuvastatin. Eur J Clin Pharmacol. 2002;58:527–31.
Ramsey LB, Johnson SG, Caudle KE, Haidar CE, Voora D, Wilke RA, et al. The Clinical Pharmacogenetics Implementation Consortium guideline for SLCO1B1 and Simvastatin-Induced Myopathy: 2014 Update. Clin Pharmacol Ther. 2014;96:423–8.
Ramsey LB, Bruun GH, Yang W, Treviño LR, Vattathil S, Scheet P, et al. Rare versus common variants in pharmacogenetics: SLCO1B1 variation and methotrexate disposition. Genome Res. 2012;22:1–8.
Nies AT, Niemi M, Burk O, Winter S, Zanger UM, Stieger B, et al. Genetics is a major determinant of expression of the human hepatic uptake transporter OATP1B1, but not of OATP1B3 and OATP2B1. Genome Med. 2013;5:1.
Kitamura S, Maeda K, Wang Y, Sugiyama Y. Involvement of multiple transporters in the hepatobiliary transport of rosuvastatin. Drug Metab Dispos. 2008;36:2014–23.
Jemnitz K, Veres Z, Tugyi R, Vereczkey L. Biliary efflux transporters involved in the clearance of rosuvastatin in sandwich culture of primary rat hepatocytes. Toxicol Vitr. 2010;24:605–10.
Pfeifer ND, Bridges AS, Ferslew BC, Hardwick RN, Brouwer KLR. Hepatic basolateral efflux contributes significantly to rosuvastatin disposition II: Characterization of hepatic elimination by basolateral, biliary, and metabolic clearance pathways in rat isolated perfused liver. J Pharmacol Exp Ther. 2013;347:737–45.
Kamiyama Y, Matsubara T, Yoshinari K, Nagata K, Kamimura H, Yamazoe Y. Role of human hepatocyte nuclear factor 4alpha in the expression of drug-metabolizing enzymes and transporters in human hepatocytes assessed by use of small interfering RNA. Drug Metab Pharmacokinet. 2007;22:287–98.
Tirona RG, Leake BF, Wolkoff AW, Kim RB, Pharmacology C, TRG T, et al. Human organic anion transporting polypeptide-C (SLC21A6) is a major determinant of rifampin-mediated pregnane X receptor activation. Pharmacology. 2003;304:223–8.
Kawashima S, Kobayashi K, Takama K, Higuchi T, Furihata T, Hosokawa M, et al. Involvement of hepatocyte nuclear factor 4alpha in the different expression level between CYP2C9 and CYP2C19 in the human liver. Drug Metab Dispos. 2006;34:1012–8.
Szatmari I, Vámosi G, Brazda P, Balint BL, Benko S, Széles L, et al. Peroxisome proliferator-activated receptor gamma regulated ABCG2 expression confers cytoprotection to human dendritic cells. J Biol Chem. 2006;281:23821–3.
Li D, Zimmerman TL, Thevananther S, Lee HY, Kurie JM, Karpen SJ. Interleukin-1 beta-mediated suppression of RXR:RAR transactivation of the Ntcp promoter is JNK-dependent. J Biol Chem. 2002;277:31416–22.
Geier A, Dietrich CG, Gerloff T, Haendly J, Kullak-Ublick GA, Stieger B, et al. Regulation of basolateral organic anion transporters in ethinylestradiol-induced cholestasis in the rat. Biochim Biophys Acta–Bio. 2003;1609:87–94.
Chew S, Lim J, Singh O, Chen X, Tan E, Lee E, et al. Pharmacogenetic effects of regulatory nuclear receptors (PXR, CAR, RXR α and HNF4 α) on docetaxel disposition in Chinese nasopharyngeal cancer patients. Eur J Clin Pharm. 2014;70:155–66.
Acknowledgements
We would like to acknowledge the University of Cape Town and Medical Research Council of South Africa for funding this research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Soko, N.D., Chimusa, E., Masimirembwa, C. et al. An African-specific profile of pharmacogene variants for rosuvastatin plasma variability: limited role for SLCO1B1 c.521T>C and ABCG2 c.421A>C. Pharmacogenomics J 19, 240–248 (2019). https://doi.org/10.1038/s41397-018-0035-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41397-018-0035-3