Abstract
Background
Exposure to inflammation exacerbates injury in neonatal encephalopathy (NE). We hypothesized that brain biomarker mRNA, cytokine mRNA and microRNA differentiate inflammation (E. coli LPS), hypoxia (Hypoxia), and inflammation-sensitized hypoxia (LPS+Hypoxia) in an NE piglet model.
Methods
Sixteen piglets were randomized: (i) LPS 2 μg/kg bolus; 1 μg/kg infusion (LPS; n = 5), (ii) Saline with hypoxia (Hypoxia; n = 6), (iii) LPS commencing 4 h pre-hypoxia (LPS+Hypoxia; n = 5). Total RNA was acquired at baseline, 4 h after LPS and 1, 3, 6, 12, 24, 48 h post-insult (animals euthanized at 48 h). Quantitative PCR was performed for cytokines (IL1A, IL6, CXCL8, IL10, TNFA) and brain biomarkers (ENO2, UCHL1, S100B, GFAP, CRP, BDNF, MAPT). MicroRNA was detected using GeneChip (Affymetrix) microarrays. Fold changes from baseline were compared between groups and correlated with cell death (TUNEL) at 48 h.
Results
Within 6 h post-insult, we observed increased IL1A, CXCL8, CCL2 and ENO2 mRNA in LPS+Hypoxia and LPS compared to Hypoxia. IL10 mRNA differentiated all groups. Four microRNAs differentiated LPS+Hypoxia and Hypoxia: hsa-miR-23a, 27a, 31-5p, 193-5p. Cell death correlated with TNFA (R = 0.69; p < 0.01) at 1–3 h and ENO2 (R = −0.69; p = 0.01) at 48 h.
Conclusions
mRNA and miRNA differentiated hypoxia from inflammation-sensitized hypoxia within 6 h in a piglet model. This information may inform human studies to enable triage for tailored neuroprotection in NE.
Impact
-
Early stratification of infants with neonatal encephalopathy is key to providing tailored neuroprotection.
-
IL1A, CXCL8, IL10, CCL2 and NSE mRNA are promising biomarkers of inflammation-sensitized hypoxia.
-
IL10 mRNA levels differentiated all three pathological states; fold changes from baseline was the highest in LPS+Hypoxia animals, followed by LPS and Hypoxia at 6 h.
-
miR-23, -27, -31-5p and -193-5p were significantly upregulated within 6 h of a hypoxia insult.
-
Functional analysis highlighted the diverse roles of miRNA in cellular processes.
Similar content being viewed by others
Introduction
Intrapartum-related neonatal encephalopathy (NE) is a significant global health burden and the third leading cause of death in children aged <5 years.1 NE has a complex and multifactorial aetiology; over the past decade, preclinical2 and clinical3 studies suggest that co-existing infection and inflammation with hypoxia–ischaemia (HI) exacerbates brain injury. A strong association exists between foetal infection/inflammation (e.g. chorioamnionitis, funisitis), perinatal brain damage and neurodisability.4
Therapeutic hypothermia (HT) initiated within 6 h of birth is now standard care for term NE in intensive care settings; cooling is only partially protective with approximately 50% of babies with adverse outcomes.5 This may be attributed, in part, to co-existing inflammation sensitization. Indeed, in a preclinical neonatal rodent study, HT was ineffective in inflammation-sensitized HI6; and in a small prospective study of placental histology relative to magnetic resonance imaging (MRI), HT was less protective in babies whose placentas showed chorioamnionitis.7 Furthermore, immune suppression associated with HT may be deleterious in infants with inflammation-sensitized brain injury.8 Given these data, the key to optimizing long-term outcomes in NE may lie in identifying biomarkers that can detect infants with prior inflammatory exposure for tailored therapeutic strategies.
A meta-analysis of brain injury biomarkers in NE identified pro-inflammatory interleukins (ILs) IL1β and IL6 and cerebrospinal fluid (CSF) neuron-specific enolase (NSE), a glycolytic enzyme found in mature neurons, as indicators of poor prognosis.9 Several other biomarkers have been investigated as indicators of injury severity and prognosis. Glial fibrillary acidic protein (GFAP), a cytoskeletal filament protein in astrocytes, has been used to stratify injury severity in NE in umbilical blood samples10 and in CSF.11 Brain-derived neurotrophic factor (BDNF) has been correlated with clinical severity of NE and poor neurological outcomes.12 Tau, a microtubule-associated protein located in axons, has been isolated in CSF in traumatic brain injuries.13 Elevated S100B, a calcium-binding protein found in glial cells, has been observed in infants with NE compared to controls in both urine and serum.14 Raised serum ubiquitin carboxyl-terminal hydrolase L1 (UCHL1), a neuron-specific enzyme, was associated with 10 min Apgar scores and abnormal MRI findings in a pilot study of infants with NE.15
More recently, micro-ribonucleic acids (miRNAs) have generated significant interest as biomarkers of neurological disease given their relative abundance in the central nervous system16 and stability in serum. These small non-coding RNAs regulate gene expression by binding to the 3’-untranslated region of a target messenger RNA (mRNA), triggering degradation or inhibiting protein translation. Several miRNA have been associated with NE, including miR-374a (piglet, human), miR-210 (piglet, rodents) and miR-376c (human).17,18,19,20
We have previously shown that exposure to Escherichia coli lipopolysaccharide (LPS) administered 4 h prior to hypoxia in newborn piglets increased mortality and exacerbated neuronal cell death beyond that observed by inflammation or hypoxia alone.21 In this current study, our aim was to evaluate gene transcript expression for several brain-specific biomarkers, cytokines and miRNA in serial blood samples taken over 48 h following hypoxia (Hypoxia), inflammation (LPS) and combined hypoxia and inflammation (LPS+Hypoxia). We hypothesized that (i) specific gene transcripts and miRNA can discriminate between inflammation sensitization with and without hypoxia; and (ii) specific gene transcripts are associated with total and regional neuronal death.
Materials and methods
The study was conducted according to UK Home Office Guidelines [Animals (Scientific Procedures) Act, 1986] and complies with the ARRIVE guidelines. The study was approved by the Ethics Committee of University College London.
Animal experiments and surgical preparation
Large white piglets (male) aged <36 h were sedated with intramuscular midazolam (150 µg) and anaesthetized with isoflurane (2–3% v/v), remaining insentient throughout experimentation. Animals were mechanically ventilated (SLE 2000, Surrey, UK) via tracheostomy and settings were adjusted to maintain partial pressure of oxygen (PaO2) and carbon dioxide (PaCO2) at 8–13 and 4.5–6.5 kPa, respectively. An umbilical venous catheter (Vygon, UK) was inserted for infusions, including maintenance 10% dextrose (60 mL/kg/day; 40 mL/kg/day post-insult), fentanyl (3 μg/kg/h) and antibiotics (benzylpenicillin 50 mg/kg BD; gentamicin 4 mg/kg OD). An umbilical arterial catheter (Vygon) was inserted for continuous monitoring of heart rate (HR) and mean arterial blood pressure (MABP) and blood sampling (pH, PaO2, PaCO2, base excess (BE), electrolytes, glucose and lactate; Abbot Laboratories, UK). A peripherally inserted central venous catheter (Vygon) was inserted for LPS or saline infusions. Post-surgery serum lactate was measured to monitor intraoperative hypoxia/hypoperfusion. Hypotension was managed by fluid boluses (0.9% saline 10 mL/kg) and inotropes; dopamine 5–25 μg/kg/min, dobutamine 5–20 μg/kg/min, noradrenaline 0.1–1 μg/kg/min and adrenaline 0.1–1 μg/kg/min were added sequentially as per neonatal guidelines. Heparinized saline (0.9% with 1 IU/mL heparin) was infused through the umbilical arterial line.
All piglets received continuous physiological monitoring (SA instruments) and intensive care support. Complications (hyperkalaemia, metabolic acidosis, seizures) were treated in accordance with neonatal guidelines. Persistent metabolic acidosis (BE > −10) was treated with sodium bicarbonate (8.4% w/v). Rectal temperature was maintained in the normothermic range (38.0–39.0 °C) using a thermoregulatory water mattress (Tecotherm).
Experimental groups
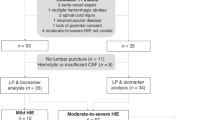
Following surgery and baseline observation, piglets were randomized by computer-generated sequence to (i) LPS, n = 5 (LPS 2 μg/kg bolus; 1 μg/kg infusion for 52 h); (ii) Hypoxia, n = 6 (saline with hypoxia); and (iii) LPS+Hypoxia, n = 5 (same dose LPS commencing 4 h prior to hypoxia) (Fig. 1).
Inflammation sensitization
E. coli LPS was used to simulate endotoxemia associated with Gram-negative sepsis. LPS and LPS+Hypoxia animals received a 2 μg/kg LPS bolus and 1 μg/kg infusion started at 4 h pre-hypoxia, modelling an ongoing infective process commencing in the hours prior to birth.
Hypoxia insult
Piglets in the Hypoxia and LPS+Hypoxia groups underwent a global hypoxia insult. Inspired oxygen (FiO2) was reduced from 21% stepwise to 4% over 3 min and held for 10 min. This was followed by a further 10–30 min of FiO2 6–12% titrated to maintain moderate hypotension (target MABP 26–30 mm Hg). Insult length was guided by real-time measurements of the duration of isoelectric electroencephalogram (EEG; <5 μV) and hypotension (MABP < 30 mm Hg). The insult was terminated early if cardiac arrest appeared imminent as indicated by profound hypotension or bradycardia.
Blood collection
Arterial bloods were taken at baseline, 4 h after the LPS bolus, immediately following insult (t = 0) and at 1, 3, 6, 12, 24 and 48 h time points post-hypoxia; 0.5 mL of whole blood was taken for gene expression analysis and mixed with acid phenol before being stored at −80 °C in cryovials for later RNA extraction.
Brain histology
Fifty-two hours following administration of an LPS bolus (48 h after hypoxia in the LPS+Hypoxia and Hypoxia groups), piglets were euthanized using intravenous pentobarbital and organs were fixed through transcardial perfusion with cold phosphate-buffered saline, followed by 4% paraformaldehyde in phosphate-buffered saline. The brain was dissected out and post-fixed at 4 °C in 4% paraformaldehyde for 7 days. Coronal slices (5-mm thick) of the right hemisphere, starting anterior to the optic chiasm, were embedded in paraffin wax and sectioned (5-µm thick). To assess cell death, sections were stained for nuclear DNA fragmentation using histochemistry with terminal deoxynucleotidyl transferase dUTP nick end labelling (TUNEL). Two sections were taken for each animal, one through the hippocampus (R1) and another 5 mm anterior (R0). Both sections were assessed for each immunohistochemical stain as described in Martinello et al.21.
RNA extraction
Total RNA was extracted from whole blood using the mirVANA miRNA Isolation Kit (Thermo Fisher Scientific, UK) in accordance with the manufacturer’s guidelines. RNA quantity was measured (ng/mL) using the Qubit RNA High Sensitivity (HS) Assay Kit (Thermo Fisher Scientific, UK). RNA was converted to complementary DNA (cDNA) using a high-capacity reverse transcription kit (SuperscriptTM VILOTM, Thermo Fisher Scientific, UK), according to the manufacturer’s guidelines; including control samples without reverse transcriptase and without sample. Total RNA samples were then aliquoted for mRNA and miRNA analysis.
mRNA amplification and analysis
Real-time quantitative PCR (qRT-PCR) was used to amplify the cDNA sequence of interest using porcine-specific Taqman probes for IL1α (IL1A), IL6 (IL6), IL8 (CXCL8), IL10 (IL10), tumour necrosis factor α (TNFA), chemokine CCL2/MCP2 (CCL2), brain-derived neurotrophic factor (BDNF), glial fibrillary acidic protein (GFAP), ubiquitin carboxyl-terminal hydrolase L1 (UCHL1), S100B (S100B), microtubule-associated protein Tau (MAPT) and neuron-specific enolase (ENO2). Three endogenous references genes were included to allow standardization: glyceraldehyde-3-phosphate dehydrogenase (GAPDH), tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta (YWHAZ), and peptidylprolyl isomerase A (PPIA).22,23,24 Target gene descriptions, function and assay ID are detailed in Table 1. Reference gene expression was not significantly different between the study groups. Customized 96-well plates (Thermo Fisher Scientific, UK) were pre-loaded with master mix (DNA polymerase, nucleotide triphosphates, buffers) and relevant primers. Samples were analyzed in triplicate. Relative quantification of gene expression was calculated using the ΔΔCT method; expression at specific time points were compared to baseline samples across all the study groups.
miRNA microarray analysis
Total RNA samples were processed by UK Bioinformatics Ltd. In brief, RNA was labelled using the FlashTagTM Biotin HSR Labelling Kit (Thermo Fisher Scientific, UK) and hybridized to GeneChip miRNA 4.0 microarrays (Affymetrix). Microarrays were preloaded to interrogate all mature miRNA sequences in miRBase, Release 20. Analysis was performed using the Expression ConsoleTM software and Transcriptome Analysis Console. Fluorescent signals generated by specific miRNA were compared to baseline values from all the groups. Results were filtered to identify statistically significant fold changes from baseline (false discovery rate (FDR) p < 0.1). Cross-species duplicates with identical sequences were removed, and the remaining miRNA were evaluated for significance (p < 0.05) at specific time points between the groups.
Gene ontology (GO) analysis
GO analysis was performed on all miRNA with significant changes from baseline to assess potential biological functions. Accession numbers of equivalent miRNA in humans were obtained from miRBase and used to build a network with previously experimentally validated miRNA–mRNA target interactions in miRTarBase version 6.1 with the CyTargetLinker plug-in in Cytoscape version 3.2.
GO analysis for molecular function, biological process, immune system, cellular component and Kyoto Encyclopedia of Genes and Genomes pathways in human species were conducted with ClueGO and Bonferroni correction for multiple testing (p < 0.01). A summarizing figure was constructed for specific terms with >5 genes/term and p < 0.01 after fusion of related terms.
Statistics
Statistical analysis was performed using IBM SPSS Statistics (v22.0), IBM Corp, Armonk, NY and JMP® (v14), SAS Institute Inc., Cary, NC, 1989–2019. Physiological data were analyzed using analysis of variance (ANOVA; Tukey–Kramer multiple comparison correction); pairwise comparisons were made using T test or Mann–Whitney U test as appropriate. Comparisons were made between groups by calculating least mean square differences and performing an ANOVA. miRNA data were log10 transformed to account for negative fold changes. Pairwise correlation (R2) was undertaken between mRNA/miRNA levels and TUNEL counts (log10 count/mm2) and mean amplitude-integrated EEG (aEEG) scores. In the event of a cardiac arrest during experimentation, all mRNA/miRNA data already acquired were included in the analysis.
Results
Baseline characteristics and physiological parameters
Sixteen large white male piglets aged <36 h were studied. Mean piglet weight was similar between groups (1960 g, range 1650–2100 g; p = 0.216). Baseline physiological parameters (HR, MBP, temperature) were within acceptable limits (Table 2). Baseline blood gases were more alkalotic in animals in the Hypoxia group. Significant differences in other parameters (core temperature, HR, MABP, BE, lactate) were within normal limits.
Hypoxia insult
Insult severity as determined by duration of hypoxia, hypotension (MABP < 27 mmHg), isoelectric EEG and area under the curve (AUC) FiO2 was similar between groups (Table 3). There was a trend towards shorter duration of EEG < 5 µV during insult in the LPS+Hypoxia group compared to Hypoxia (p = 0.07). End of insult blood gases were similar between groups.
Survival and clinical illness severity
As reported in Martinello et al.,21 mortality was significantly increased in LPS+Hypoxia; 3 out of 5 LPS+Hypoxia piglets died compared to none in the other groups (p = 0.022). One piglet died within minutes of insult cessation; two piglets arrested at approximately 24 h (mRNA data were acquired up to 24 h). Two piglets (one LPS, one Hypoxia) required resuscitation following mechanical airway obstruction and survived to experiment completion.
Both LPS and LPS+Hypoxia animals required saline boluses and multiple inotropes to maintain haemodynamic stability; however, there was no significant overall difference between the groups (Table 4). Hypoxia and LPS+Hypoxia animals required higher doses of dopamine, dobutamine, adrenaline and saline than LPS animals (p ≤ 0.046).
Gene expression for cytokines and chemokines
Total RNA was acquired for all LPS and Hypoxia animals. In LPS+Hypoxia, RNA was obtained in 4 out of 5 animals until 24 h and 2 out of 5 animals at 48 h (Supplementary File S1).
Comparisons of gene transcript levels at specific time points identified that pro-inflammatory (IL1Α), chemotactic (CXCL8, CCL2) and anti-inflammatory (IL10) mRNA were significantly upregulated in LPS+Hypoxia and LPS compared to Hypoxia within 6 h post-insult (p < 0.01; Fig. 2). IL10 mRNA levels differentiated all three pathological states; fold changes from baseline were highest in LPS+Hypoxia, followed by LPS and Hypoxia at 6 h (p < 0.033). Elevated ILA and IL10 mRNA levels persisted in animals receiving LPS+Hypoxia, compared to LPS and Hypoxia. IL6 mRNA levels, thought to have both pro- and anti-inflammatory activities, was higher at 3 h in LPS+Hypoxia compared to LPS (p = 0.01) with a trend to an increase compared to Hypoxia animals (p = 0.06) and a second peak at 24 h post-insult (p = 0.03). CXCL8 mRNA also demonstrated a biphasic response to LPS+Hypoxia, peaking at 3–6 h and 24 h post-insult (p < 0.01). Pro-inflammatory TNFΑ mRNA decreased in the first 6 h in all the study groups; however, it increased at 24 h post-insult in LPS+Hypoxia and LPS compared to Hypoxia (p < 0.01). Complete mRNA data are listed in Supplementary File S1.
Gene expression for brain-specific biomarkers
ENO2 mRNA (neuron-specific enolase) significantly increased at 1 h post-insult in LPS+Hypoxia and LPS compared to Hypoxia (p = 0.04 and p = 0.01, respectively). The expression of this transcript was also significantly higher at 3 h in LPS+Hypoxia compared to LPS (p = 0.05; Fig. 3).
a–e ENO2 (b) was the only brain biomarker to differentiate between Hypoxia and LPS+Hypoxia within 6 h. BDNF (a) and MAPT (c) mRNA were significantly upregulated in LPS+Hypoxia at 24 h post-insult. Expression of GFAP and CRP mRNA was insufficient for analysis. BDNF brain-derived neurotrophic factor, ENO2 enolase 2, MAPT microtubule-associated protein Tau, S100B S100B protein, UCHL1 ubiquitin carboxyl-terminal hydrolase L1, GFAP glial fibrillary acidic protein, CRP C-reactive protein.
MAPT mRNA (encoding Tau protein) increased in LPS at 6 and 48 h (p = 0.04) and LPS+Hypoxia at 24 h (p < 0.04) compared to Hypoxia. BDNF mRNA (neurotrophic factor) was higher in LPS+Hypoxia compared to Hypoxia from 3 h post-insult, reaching statistical significance at 24 h (p = 0.03).
UCHL1 mRNA (a neuron-specific enzyme) did not distinguish between the groups at any time points. S100B mRNA, predominantly found in glial cells, varied significantly between the groups at baseline and remained elevated in animals receiving LPS. GFAP mRNA (cytoskeletal filament protein) and CRP (inflammatory biomarker) were undetectable in several samples across all the study groups and insufficient for analysis.
Expression of miRNA
In total, 4929 miRNAs were analysed; we identified 22 miRNAs with significantly altered expression from baseline (FDR p value < 0.1; see Supplementary File S2). Significant changes from baseline levels were seen in 12 miRNAs in LPS (2 up, 10 down), 11 miRNAs in Hypoxia (all down) and 7 miRNAs in LPS+Hypoxia (2 up, 5 down).
Out of the 22 miRNA, we identified 8 promising candidates that discriminated between the study groups at early (<6 h) and late (24 h) time points. In the first 6 h post-insult, hsa-mir-23a-5p and hsa-mir-27a-5p levels increased significantly in LPS and LPS+Hypoxia compared to Hypoxia at 1, 3 and 6 h (all p < 0.01). Also, hsa-mir-31-5p and hsa-mir-193-5p levels were significantly lower in LPS+Hypoxia and LPS compared to Hypoxia immediately post-insult (p < 0.01), though this difference was no longer significant by 1 h (p = 0.06) (Fig. 4a–d). There were 4 miRNAs demonstrating differential expression between the groups at 24 h: hsa-mir-181c-3p, hsa-mir-330-3p, hsa-mir-182-5p, and hsa-mir-150-5p (Fig. 4e–h).
Biomarkers of injury severity
Candidate biomarkers were assessed as markers of injury severity by correlating fold changes with overall TUNEL counts (cell death) and aEEG scores.
We observed a significant positive correlation between overall TUNEL-positive cell counts and TNFA (R = 0.69, p < 0.01) and S100B (R = 0.53, p = 0.04) mRNA at 1 h post-insult and a negative correlation in ENO2 mRNA at 48 h (R = −0.69, p = 0.01). Although IL10 mRNA levels appeared to coincide with injury severity, we did not find a statistically significant correlation with cell death.
We identified 9 out of 22 significantly altered miRNAs that correlated with TUNEL-positive counts within 6 h of insult: ssc-mir-31 (R = 0.59, p = 0.027), ssc-mir-199a-5p (R = 0.79, p = 0.001) and hsa-mir-214-5p (R = 0.66, p = 0.01) at 1 h post-insult; hsa-mir-150-5p (R = 0.63, p = 0.02) at 3 h post-insult; ssc-mir-4334-3p (R = 0.79, p = <0.01) and hsa-mir-181c-3p (R = 0.73, p < 0.01) at 6 h post-insult. Notably, hsa-mir-193a-5p and ssc-mir-99b were strongly associated with cell death at 1, 3 and 6 h post-insult (R > 0.70, p < 0.01 at 6 h).
There was no significant correlation between aEEG scores and mRNA/miRNA levels (data analysis included in Supplementary File S3).
miRNA and mRNA networks
There were several unique and overlapping predicted mRNA targets associated with the miRNA isolated at 12 h in LPS, Hypoxia and LPS+Hypoxia (Fig. 5). Nucleic acid metabolism, transcription and translation were affected in all the three groups, though greatest in the Hypoxia group. HI is known to affect genomic integrity, triggering activation of the repair enzyme poly (ADP-ribose) polymerase-1, found only in the Hypoxia group. While repair of DNA is beneficial, overactivation signals irreparable injury and triggers apoptosis. All insults affected cell-cycle control proteins, particularly p53, which lead to enrichment of cancer-related pathways. Genes related to mitochondrial membrane permeabilization were enriched in all the study groups. Pathways downregulating oxidative stress-induced cell death was observed in LPS+Hypoxia only.
GO analysis also highlighted several differences in the predicted biological pathways. The Hypoxia group-enriched pathways related to brain development (forebrain development, regulation of neurogenesis, neurotrophin signalling pathways), cellular components (spindles, chromosomes) and protein activity (protein deacylation, serine/threonine kinase activity). Organ development pathways appeared more enriched in LPS+Hypoxia and LPS, compared to Hypoxia animals, suggesting a more systemic nature to the injury mechanism. Interestingly, the signalling pathways for pro-inflammatory transcription factor nuclear factor-kappa B (NF-κB) were enriched in the LPS group but not in the LPS+Hypoxia group (Fig. 6a–c). Full GO data are included in Supplementary File S4.
Discussion
Using a piglet model of inflammation-sensitized hypoxia, we present the analysis of serial serum inflammatory and brain-specific biomarkers that discriminate between hypoxia associated with and without inflammation within 6 h of the hypoxia insult.
Cytokines
Several studies have observed an association between raised pro-inflammatory cytokines and adverse neurological sequelae in infants with NE.10 Elevated IL1α, 6, 8 and TNFα in the first 24 h correlate with abnormal MRI25 and poor neurodevelopmental outcomes at 12–24 months.10,25 In our study, animals receiving an inflammatory insult had significantly higher IL1A, CXCL8, CCL2 and IL10 mRNA within 3 h of insult compared to hypoxia injury. These biomarkers may help differentiate the predominate pathological process in infants with NE.
Cytokines and chemokines are not exclusively pro- or anti-inflammatory; for example, IL6 increases vascular permeability and causes cerebral oedema at high concentrations26; however, it also inhibits TNFα and IL1 and promotes nerve growth factor secretion.27 IL6 has been reported as a poor prognostic marker in some10,28,29 but not all studies.30 We observed a twofold increase in IL6 mRNA in Hypoxia and eightfold in LPS+Hypoxia animals within 3 h of injury, reflecting the increased injury associated with dual pathology. Interestingly, we noted a secondary peak in IL6 and CXCL8 (encoding IL8) mRNA at 24 h in LPS+Hypoxia. A similar biphasic profile was observed in HT-treated infants with good outcomes,28 suggesting this increase was not necessarily pathological. Persistent elevation of IL6, however, was associated with a poor prognosis. The delayed rise in IL6 and IL8 may represent activation of reparative processes, the success of which determines whether these cytokines subsequently fall or remain elevated. In our study, this biphasic pattern was observed in LPS+Hypoxia only, the group with most severe injury. Owing to the limited data at 48 h, it remains unclear whether these cytokines would have remained elevated had more animals survived.
The most promising differentiating biomarker among the cytokines was IL10 mRNA, distinguishing between all three pathological states; transcripts were the highest in LPS+Hypoxia and the lowest in Hypoxia at 6 h post-insult. IL10 is predominately anti-inflammatory31 and has shown neuroprotective benefit in small animal models of NE. Raised IL10, however, is associated with adverse outcomes in NE28,32 and sepsis,33 possibly reflecting an exaggerated response to severe infection/inflammation. Although its precise role in the evolution of injury is unclear, it remains a useful marker differentiating inflammation from hypoxia injury.
Brain biomarker
Disruption to the blood–brain barrier and leakage of brain-specific proteins into the extracellular space permits detection of these biomarkers in serum,34,35 urine36 and CSF.9,37 ENO2 mRNA was the only brain biomarker significantly altered in LPS+Hypoxia compared to Hypoxia within 6 hours post-insult. The ENO2 gene encodes for NSE, a glycolytic enzyme predominately found in neurons. Elevated NSE in serum38 and CSF37 have been associated with increased mortality and adverse outcomes at 12 months in infants with NE. Though predominately found in neurons, NSE is also expressed in red blood cells; serum levels may therefore overestimate brain injury, particularly in the presence of haemolysis. Importantly, ENO2 mRNA differentiated injury subtypes within the 6-h therapeutic window for HT, providing an opportunity to initiate alternative strategies.
Other brain-specific biomarkers investigated were not as discriminative. MAPT (Tau) mRNA levels appeared higher in LPS+Hypoxia compared to Hypoxia throughout the experiment, though only reached statistical significance at 24 h. Similarly, expression of BDNF, a neurotrophin secreted by neurons and astrocytes, was elevated at 24 h only in the LPS+Hypoxia group. The mRNA for UCHL1, a gene encoding for a cysteine protease expressed in neurons, was increased in all the groups; however, it did not differentiate between different injury subtypes. Surprisingly, GFAP mRNA, a cytoskeletal protein in astrocytes, was undetectable in several samples despite the extensive cell death seen in some animals.
Markers of injury severity
Combining data from all animals, we examined whether fold changes in cytokines or brain biomarkers were associated with TUNEL-positive cells (cell death). TNFA mRNA was the only cytokine that correlated with overall TUNEL counts at 1 and 3 h, indicating that it may be an early marker of injury severity. ENO2 mRNA negatively correlated with cell death at 48 h post-insult. Our group previously identified a significant correlation between serum IL1β, IL10 and CSF TNFα and MRS biomarkers (Lac/NAA) and IL1β and IL8 with TUNEL-positive cells in the thalamus in a piglet model of NE.30 Gene expression of these cytokines, however, was not associated with regional or overall cell death. Patterns of aEEG are useful prognostic indicators in NE39; we, however, did not find significant correlation with any cytokine or brain biomarker transcript levels.
miRNA biomarkers
miRNAs are non-coding sequences of RNA that regulate gene expression and play an essential role in early neurological development.40 We identified four promising candidates as early differentiating biomarkers of infection-sensitized hypoxia: hsa-mir-23a, hsa-mir-27a, hsa-mir-31a-5p, and hsa-mir-193-5p. Both miR-23 and mir-27, in combination with miR-24, play an important role in T cell activation and differentiation. Mir-31, analogous to mir-31a-5p, regulates transcription factors, including hypoxia-inducible-factor 1α (HIF1α) and NF-κB. Both hsa-mir-31 and hsa-mir-193-5p correlated with neuronal cell death 1 h post-insult, therefore these may be indicators of injury severity.
Hsa-mir-181c-3p, hsa-mir-330-3p, hsa-mir-182-p and hsa-mir-150-5p were downregulated in all the groups, particularly in LPS and LPS+Hypoxia compared to Hypoxia animals at 24 h. These miRNAs were significantly induced following LPS and may be useful in identifying ongoing infective or inflammatory pathologies. mir-150-5p is highly expressed in mature B cells and T cells, playing a critical role in immune cell differentiation. mir-150-5p has also been identified as a prognostic marker following acute ischaemic stroke,41 consistent with the positive correlation with overall cell death in this study. mir-181c-3p has previously been linked to the hypoxic response,42 downstream of HIF1α.43 We identified a positive correlation between this miRNA and neuronal cell death, suggesting a potential role in the pathogenesis of neurological injury, which needs further study.
Several miRNAs have been identified to play a key role in HI and neuroinflammation. The most notable being miR-210, referred to as a master hypoxamiR and consistently altered in adult rodent models of ischaemic stroke.19,44 Serum miR-210 was increased in pregnancies complicated by foetal growth restriction, a condition associated with chronic foetal hypoxia.45 miRNA may influence several genes simultaneously and are therefore well placed as regulators of inflammation. miR-155 has potent pro-inflammatory properties, including the downregulation of suppressor of cytokine signalling 1 protein and increasing cytokine and nitric oxide production.46 miR-146a is anti-inflammatory, downregulating the TNF receptor-associated factor 6 and interleukin-1 receptor-associated kinase 1 adaptor molecules and thus inhibiting Toll-like receptor 4 activation. Interestingly, we did not observe significant changes in these miRNAs in any groups in this study.
Functional analysis of miRNAs highlighted several key pathophysiological processes, including cell cycle, signal transduction, DNA damage, transcription/translation and apoptosis. Genes related to mitochondrial membrane permeability were enriched in all the groups, confirming mitochondrial injury as one of the hallmarks of secondary energy failure in NE. Downregulation of oxidative stress-induced cell death pathways was only seen in LPS+Hypoxia and may represent activation of compensatory mechanisms.
Timing of injury
Identifying the timing of perinatal brain injury is a significant challenge in clinical practice and has significant implications on both prognosis and efficacy of treatment. Our model identified combinations of biomarkers that reached peak levels at different time points post-HI: ENO2 and CCL2 at 1–3 h, ILA and IL10 by 6 h, and TNFA and MAPT at 24 h. However, these data are from a model with precise timings of inflammation and hypoxia, and application in the clinical setting is much more complex.
Limitations
The main limitation of the study was the high mortality in LPS+Hypoxia, reducing available data at 48 h; we were, however, able to obtain mRNA/miRNA for 15 out of 16 animals up to 24 h. The early mortality in LPS+Hypoxia limits the interpretation of results at 48 h. These animals represent the most severe spectrum of injury, and therefore our results may underestimate cytokine and brain biomarker mRNA levels at this time point.
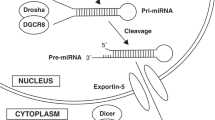
Animals in this study were not cooled. As HT may be less effective in inflammation-sensitized hypoxia, we focussed on biomarkers during normothermia within the 6-h treatment window to facilitate decision-making to initiate alternative neuroprotective strategies. HT may influence RNA dynamics through the induction of cold-inducible binding proteins, decreasing inflammatory cytokine production47 and attenuating caspase-dependent apoptosis.48 HT also alters miRNA regulation; miR-874 and miR-451 were significantly increased in normothermia compared to HT-treated adult rodents 7 h post-traumatic brain injury.49 HT induces the glycine-rich RNA-binding protein (RBM3), disinhibiting Dicer, the enzyme responsible for cleaving precursor pre-miRNA into its mature counterpart.50 Further study in HT-treated animals is warranted.
There are also limitations when inferring injury mechanisms using gene expression, particularly as proteins are subject to posttranslational regulation and may not correlate with their respective mRNA transcripts.51
This study raises more questions that could be investigated in future work. The following are important areas to study: evaluation of the influence of timing of LPS administration on cytokine expression and outcome, exploration of brain tissue mRNA expression to establish the specific cell lines involved, correlation of gene expression with protein levels, and exploration of gender-specific differences.
Conclusions
Using a large animal model of inflammation-sensitized hypoxia, we investigated an extensive panel of biomarkers at clinically relevant time points. Within 6 h of insult, IL1Α, CXCL8, IL10, CCL2 and ENO2 mRNA increased in animals receiving LPS+Hypoxia and LPS compared to Hypoxia. IL10 mRNA differentiated all three pathological states within this timeframe. These biomarkers may inform the triage of tailored treatments for infants with NE. This study also demonstrates the feasibility and clinical utility of mRNA-based biomarkers, supporting the development of gene expression testing as point-of-care diagnostics. These data require validation in the clinical setting and may facilitate tailored neuroprotective strategies to improve long-term outcomes.
References
Lee, A. C. C. C. et al. Intrapartum-related neonatal encephalopathy incidence and impairment at regional and global levels for 2010 with trends from 1990. Pediatr. Res. 74, 50–72 (2013).
Eklind, S. et al. Bacterial endotoxin sensitizes the immature brain to hypoxic-ischaemic injury. Eur. J. Neurosci. 13, 1101–1106 (2001).
Nelson, K. B. & Willoughby, R. E. Infection, inflammation and the risk of cerebral palsy. Curr. Opin. Neurol. 13, 133–139 (2000).
Tann, C. J. et al. Perinatal risk factors for neonatal encephalopathy: an unmatched case-control study. Arch. Dis. Child. Fetal Neonatal Ed. 103, F250–F256 (2018).
Jacobs, S. E. et al. Cooling for newborns with hypoxic ischaemic encephalopathy. Cochrane Database Syst. Rev. 2013, CD003311 (2013).
Osredkar, D. et al. Hypothermia is not neuroprotective after infection-sensitized neonatal hypoxic–ischemic brain injury. Resuscitation 85, 567–572 (2014).
Impey, L. W. & Greenwood, C. E. B. R. The relationship between intrapartum maternal fever and neonatal acidosis as risk factors for neonatal encephalopathy. Am. J. Obstet. Gynecol. 198, 49.e1 (2008).
Jenkins, D. D. et al. Altered circulating leukocytes and their chemokines in a clinical trial of therapeutic hypothermia for neonatal hypoxic ischemic encephalopathy. Pediatr. Crit. Care Med. 14, 786–795 (2013).
Ramaswamy, V. et al. Systematic review of biomarkers of brain injury in term neonatal encephalopathy. Pediatr. Neurol. 40, 215–226 (2009).
Chalak, L. F. et al. Biomarkers for severity of neonatal hypoxic-ischemic encephalopathy and outcomes in newborns receiving hypothermia therapy. J. Pediatr. 164, 468.e1–474.e1 (2014).
Blennow, M., Hagberg, H. & Rosengren, L. Glial fibrillary acidic protein in the cerebrospinal fluid: a possible indicator of prognosis in full-term asphyxiated newborn infants? Pediatr. Res. 37, 260–264 (1995).
Imam, S. S., Gad, G. I., Atef, S. H. & Shawky, M. A. Cord blood brain derived neurotrophic factor: diagnostic and prognostic marker in fullterm newborns with perinatal asphyxia. Pak. J. Biol. Sci. 12, 1498–1504 (2009).
Zemlan, F. P. et al. C-tau biomarker of neuronal damage in severe brain injured patients: association with elevated intracranial pressure and clinical outcome. Brain Res. 947, 131–139 (2002).
Risso, F. M. et al. Urine S100 BB and A1B dimers are valuable predictors of adverse outcome in full-term asphyxiated infants. Acta Paediatr. Int. J. Paediatr. 102, 467–472 (2013).
Douglas-Escobar, M. et al. A pilot study of novel biomarkers in neonates with hypoxic-ischemic encephalopathy. Pediatr. Res. 68, 531–536 (2010).
Yang, Y. Y. et al. Non-coding RNAs in human disease. J. Neuropathol. Exp. Neurol. 20, 77–103 (2013).
Looney, A.-M. et al. Downregulation of umbilical cord blood levels of miR-374a in neonatal hypoxic ischemic encephalopathy. J. Pediatr. 167, 269–273.e2 (2015).
Garberg, H. T. et al. Temporal profile of circulating microRNAs after global hypoxia-ischemia in newborn piglets. Neonatology 111, 133–139 (2017).
Qiu, J. et al. Neuroprotective effects of microRNA-210 on hypoxic-ischemic encephalopathy. Biomed. Res. Int. 2013, 350419 (2013).
O’Sullivan, M. P. et al. Validation of altered umbilical cord blood microRNA expression in neonatal hypoxic-ischemic encephalopathy. JAMA Neurol. 76, 333 (2019).
Martinello, K. A. et al. Acute LPS sensitization and continuous infusion exacerbates hypoxic brain injury in a piglet model of neonatal encephalopathy. Sci. Rep. 9, 10184 (2019).
Uddin, M. J. et al. Age-related changes in relative expression stability of commonly used housekeeping genes in selected porcine tissues. BMC Res. Notes 4, 441 (2011).
Wang, J. et al. Selection of reference genes for gene expression studies in porcine whole blood and peripheral blood mononuclear cells under polyinosinic:polycytidylic acid stimulation. Asian-Australas. J. Anim. Sci. 27, 471–478 (2014).
Robertson, N. J. et al. Melatonin augments hypothermic neuroprotection in a perinatal asphyxia model. Brain 136, 90–105 (2013).
O’Hare, F. M. et al. Serial cytokine alterations and abnormal neuroimaging in newborn infants with encephalopathy. Acta Paediatr. 106, 561–567 (2017).
Lv, H. et al. Neonatal hypoxic ischemic encephalopathy-related biomarkers in serum and cerebrospinal fluid. Clin. Chim. Acta 450, 282–297 (2015).
Kossmann, T., Hans, V., Imhof, H. G., Trentz, O. & Morganti-Kossmann, M. C. Interleukin-6 released in human cerebrospinal fluid following traumatic brain injury may trigger nerve growth factor production in astrocytes. Brain Res. 713, 143–152 (1996).
Jenkins, D. D. et al. Serum cytokines in a clinical trial of hypothermia for neonatal hypoxic-ischemic encephalopathy. J. Cereb. Blood Flow Metab. 32, 1888–1896 (2012).
Chiesa, C. et al. Umbilical cord interleukin-6 levels are elevated in term neonates with perinatal asphyxia. Eur. J. Clin. Invest. 33, 352–358 (2003).
Rocha-Ferreira, E. et al. Systemic pro-inflammatory cytokine status following therapeutic hypothermia in a piglet hypoxia-ischemia model. J. Neuroinflammation 14, 44 (2017).
Garcia, J. M. et al. Role of interleukin-10 in acute brain injuries. Front. Neurol. 8, 244 (2017).
Orrock, J. E. et al. Association of brain injury and neonatal cytokine response during therapeutic hypothermia in newborns with hypoxic-ischemic encephalopathy. Pediatr. Res. 79, 742–747 (2016).
Derkx, B. et al. High levels of interleukin-10 during the initial phase of fulminant meningococcal septic shock. J. Infect. Dis. 171, 229–232 (1995).
Ennen, C. S. et al. Glial fibrillary acidic protein as a biomarker for neonatal hypoxic-ischemic encephalopathy treated with whole-body cooling. Am. J. Obstet. Gynecol. 205, 251.e1–251.e7 (2011).
Massaro, A. N. et al. Serum biomarkers of MRI brain injury in neonatal hypoxic ischemic encephalopathy treated with whole-body hypothermia. Pediatr. Crit. Care Med. 14, 310–317 (2013).
Alshweki, A., Pérez-Muñuzuri, A., López-Suárez, O., Baña, A. & Couce, M. L. Relevance of urinary S100B protein levels as a short-term prognostic biomarker in asphyxiated infants treated with hypothermia. Medicine (Baltimore) 96, e8453 (2017).
Vasiljevic, B. et al. New insights into the pathogenesis of perinatal hypoxic-ischemic brain injury. Pediatr. Int. 53, 454–462 (2011).
Çeltik, C., Acunaş, B., Öner, N. & Pala, Ö. Neuron-specific enolase as a marker of the severity and outcome of hypoxic ischemic encephalopathy. Brain Dev. 26, 398–402 (2004).
Merchant, N. & Azzopardi, D. Early predictors of outcome in infants treated with hypothermia for hypoxic-ischaemic encephalopathy. Dev. Med. Child Neurol. 57, 8–16 (2015).
Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 12, 861–874 (2011).
Scherrer, N. et al. MicroRNA 150-5p improves risk classification for mortality within 90 days after acute ischemic stroke. J. Stroke 19, 323–332 (2017).
Kulshreshtha, R. et al. A microRNA signature of hypoxia. Mol. Cell. Biol. 27, 1859–1867 (2007).
Camps, C. et al. Integrated analysis of microRNA and mRNA expression and association with HIF binding reveals the complexity of microRNA expression regulation under hypoxia. Mol. Cancer 13, 28 (2014).
Meng, Z. Y. et al. MicroRNA-210 promotes accumulation of neural precursor cells around ischemic foci after cerebral ischemia by regulating the SOCS1-STAT3-VEGF-C pathway. J. Am. Heart Assoc. 7, e005052 (2018).
Whitehead, C. L. et al. Circulating microRNAs in maternal blood as potential biomarkers for fetal hypoxia in-utero. PLoS ONE 8, e78487 (2013).
Cardoso, A. L., Guedes, J. R., Pereira de Almeida, L. & Pedroso de Lima, M. C. miR-155 modulates microglia-mediated immune response by down-regulating SOCS-1 and promoting cytokine and nitric oxide production. Immunology 135, 73–88 (2012).
Kaneko, T. & Kibayashi, K. Mild hypothermia facilitates the expression of cold-inducible RNA-binding protein and heat shock protein 70.1 in mouse brain. Brain Res. 1466, 128–136 (2012).
Wang, D. & Zhang, J. Effects of hypothermia combined with neural stem cell transplantation on recovery of neurological function in rats with spinal cord injury. Mol. Med. Rep. 11, 1759–1767 (2015).
Truettner, J. S., Alonso, O. F., Bramlett, H. M. & Dietrich, W. D. Therapeutic hypothermia alters microRNA responses to traumatic brain injury in rats. J. Cereb. Blood Flow Metab. 31, 1897–1907 (2011).
Pilotte, J., Dupont-Versteegden, E. E. & Vanderklish, P. W. Widespread regulation of miRNA biogenesis at the dicer step by the cold-inducible RNA-binding protein, RBM3. PLoS ONE 6, e28446 (2011).
Liu, Y., Beyer, A. & Aebersold, R. On the dependency of cellular protein levels on mRNA abundance. Cell 165, 535–550 (2016).
Kawata, K., Liu, C. Y., Merkel, S. F., Ramirez, S. H., Tierney, R. T., Langford, D. Blood biomarkers for brain injury: What are we measuring? Neurosci Biobehav Rev. 68, 460–473 (2016).
Acknowledgements
We thank Debbie Kraus (statistical analysis), David Chambers and Geoff Scopes (UK Bioinformatics) for their support in this study. This study was funded by the Medical Research Council (MR/M006743/1). This work was undertaken at University College London Hospitals/University College London, which received a proportion of funding from the UK Department of Health’s National Institute for Health Research Biomedical Research Center’s funding scheme.
Author information
Authors and Affiliations
Contributions
The project grant was authored by N.J.R., D.P., H.H., P.G., B.F., N.K., B.W.K., T.G.A.M.W. and C.J.T. Experiments were performed by I.L., A.A.-B., C.M., K.M. and S.R. Isolation of total RNA and qPCR was performed by I.L., A.A.-B., S.R. and C.M. Processing of RNA for miRNA detection was performed by UK Bioinformatics Ltd. Analysis of gene expression and miRNA data was undertaken by I.L., B.F. and P.G. GO analysis, interpretation and figure illustration was performed by M.B. and revised by I.L. K.M. contributed to the analysis of the physiological data and immunohistochemistry. The manuscript was drafted by I.L. and revised by N.J.R., B.F. and B.K. All authors have read and agreed to the submitted version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lingam, I., Avdic-Belltheus, A., Meehan, C. et al. Serial blood cytokine and chemokine mRNA and microRNA over 48 h are insult specific in a piglet model of inflammation-sensitized hypoxia–ischaemia. Pediatr Res 89, 464–475 (2021). https://doi.org/10.1038/s41390-020-0986-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-020-0986-3