Abstract
Background
Neuroblastoma is the second most common extracranial cancer in children. Current therapies for neuroblastoma, which use a combination of chemotherapy drugs, have limitations for high-risk subtypes and can cause significant long-term adverse effects in young patients. Therefore, a new therapy is needed. In this study, we investigated the transcription factor MXD3 as a potential therapeutic target in neuroblastoma.
Methods
MXD3 expression was analyzed in five neuroblastoma cell lines by immunocytochemistry and quantitative real-time reverse transcription PCR, and in 18 primary patient tumor samples by immunohistochemistry. We developed nanocomplexes using siRNA and superparamagnetic iron oxide nanoparticles to target MXD3 in neuroblastoma cell lines in vitro as a single-agent therapeutic and in combination with doxorubicin, vincristine, cisplatin, or maphosphamide—common drugs used in current neuroblastoma treatment.
Results
MXD3 was highly expressed in neuroblastoma cell lines and in patient tumors that had high-risk features. Neuroblastoma cells treated in vitro with the MXD3 siRNA nanocomplexes showed MXD3 protein knockdown and resulted in cell apoptosis. Furthermore, on combining MXD3 siRNA nanocomplexes with each of the four drugs, all showed additive efficacy.
Conclusion
These results indicate that MXD3 is a potential new target and that the use of MXD3 siRNA nanocomplexes is a novel therapeutic approach for neuroblastoma.
Similar content being viewed by others
Main
Neuroblastoma is a cancer of the sympathetic nervous system and the most common extracranial solid tumor in children (1). Despite improvement in outcome for patients with low- and mid-risk subtypes of neuroblastoma, the outcome for patients with high-risk subtypes remains poor, with survival rates as low as 50% (2). In addition, many survivors develop irreversible, long-term adverse effects from systemic chemotherapy and radiation (3). Therefore, there is a need for treatments that are more effective and less toxic than current therapies.
One new therapeutic approach uses siRNA to target genes that are essential for cancer survival and progression (4). Using siRNA to target specific cancer-associated genes could increase treatment efficacy and potentially replace or reduce doses of currently used chemotherapy drugs, ameliorating undesirable adverse effects. siRNA has already been used in clinical trials, for example, to target BCR-ABL for chronic myeloid leukemia and KRAS for pancreatic cancer with some success (5, 6). Therefore, there is great potential in further expanding siRNA therapeutics to other cancers, including pediatric cancers such as neuroblastoma. Several genes have been identified and studied as therapeutic targets for neuroblastoma, such as MYCN and ALK (7, 8). As monotherapy does not work for most cancers, an ideal therapy should include multiple molecular-targeted approaches. Previously, we identified the transcription factor MXD3 as a novel target for precursor B-cell acute lymphoblastic leukemia (preB ALL) (9, 10, 11). MXD3 is a member of the family of Mad proteins that interact with Max proteins (12), and is reported to have a role in medulloblastoma tumorigenesis (13). Furthermore, a public microarray data set (R2: Genomics Analysis and Visualization Platform) shows that patients with high MXD3 neuroblastoma have a significantly inferior survival rate compared with those with low MXD3 neuroblastoma (http://hgserver1.amc.nl/cgi-bin/r2/main.cgi). We hypothesized that MXD3 is important for neuroblastoma cell survival; therefore, targeting MXD3 using siRNA can be of therapeutic potential.
One of the major challenges for siRNA therapeutics is efficient siRNA delivery into tumor cells (14). Unmodified siRNA is large compared with chemotherapy drugs such as doxorubicin or vincristine, negatively charged, and cannot pass through cell membranes by itself (15). One way to solve this problem is to use a vehicle, such as nanoparticles (NPs), to facilitate intracellular delivery (14). Multiple types of NPs have been studied (16). For instance, liposomal NPs carrying chemically stabilized siRNA against PKN3 were used to treat 16 different types of advanced solid tumors such as breast, pancreatic, colon, and ovarian cancers (17). In another study, polymer NPs using a human transferrin protein-targeting ligand were used to treat prostate, gastrointestinal, and melanoma tumors by delivering siRNA against RRM2 intracellularly (18). Metal NPs have also been studied for clinical use (19). For instance, many superparamagnetic iron oxide (SPIO) NP formulations have been approved for MRI imaging and diagnostics (20). SPIO NPs have been used to develop a new strategy for treating tumors with magnetic-induced hyperthermia (21). SPIO NPs have also been studied to deliver siRNA to inhibit apoptosis in transplanted pancreatic β-cells (22). These studies show that SPIO NPs have the potential to be used as a therapeutic vehicle. In our study, we investigated SPIO NPs with a modified polyethylenimine (PEI) surface as our delivery vehicle for MXD3 siRNA.
In summary, our study identified and validated a new therapeutic target MXD3, and demonstrated a novel therapeutic approach using siRNA and SPIO NPs for neuroblastoma. The results of this study should help guide future preclinical and clinical studies to develop and validate this new strategy for neuroblastoma treatment.
Methods
Cells and Cell Cultures
The human neuroblastoma cell lines SH-SY5Y, IMR-32, SK-N-DZ, SK-N-BE, and SK-N-SH were purchased from ATCC (Manassas, VA). Cells were used within the first 25 passages. SK-N-DZ cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, ATCC), supplemented with non-essential amino acids (Thermo Fisher Scientific, Waltham, MA). SH-SY5Y and SK-N-BE cells were cultured in a 1:1 mixture of Modified Eagle Medium (MEM) and F12 medium, supplemented with non-essential amino acids, and 100 mM sodium pyruvate (all Thermo Fisher Scientific). IMR-32 and SK-N-SH cells were cultured in MEM supplemented with non-essential amino acids, and 100 mM sodium pyruvate (Thermo Fisher Scientific). All media formulations were further supplemented with 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin (all Thermo Fisher Scientific).
Immunochemistry and Fluorescence Intensity Quantification
MXD3 protein expression was evaluated by immunochemistry as previously described (10). Briefly, cells were fixed with 10% buffered formalin, smeared onto slides, and blocked with 10% FBS. Slides were incubated with anti-MXD3 monoclonal mouse antibody (Antibodies, Davis, CA) overnight at 4 °C. Incubation with secondary goat anti-mouse antibody conjugated to Alexa Fluor 488 (A488) (Thermo Fisher Scientific) was performed at room temperature for 3 h. MXD3 protein expression was quantified by measuring the fluorescent image intensity of all cells within a representative image (TIFF) and averaged to obtain the mean fluorescence intensity. Measurement and quantification was performed using ImageJ (NIH) (23). Briefly, individual cell boundaries were marked and the mean fluorescence intensity (MFI) was measured for each cell. A background signal (without cells) was subtracted from each MFI. Corrected MFI for each cell was then averaged per experiment and treatment type.
Tumor microarrays with 18 human neuroblastoma tissue samples were stained for MXD3 at the University of California, Davis (UC Davis) Cancer Center Pathology Core.
Western Blot
Frozen whole-cell pellets were prepared for western blotting as described previously (11). Briefly, cell pellets were denatured and boiled, run on a 4-20% Tris-Glycine SDS-PAGE gel, and transferred to a nitrocellulose membrane using a wet transfer system. Blots were blocked, incubated with primary antibodies overnight at 4 °C, and then incubated with secondary antibodies for 1 h at room temperature. Blots were washed and then incubated with SuperSignal West Pico Chemiluminescent kit (Thermo Fisher Scientific) according to the manufacturer’s instructions for imaging.
Reagents
siRNA molecules and NPs were purchased as described in our previous study (10). The MXD3 siRNA sense strand sequence was 5′-AUGGACUAAAAGGACCCUUTT-3′, and the antisense strand was 5′-AAGGUCCUUUAGUCCAUTT-3′. AllStars Negative siRNA (Qiagen, Hilden, Germany) was used as a negative control. Both MXD3 and control siRNAs were tagged with A488 on the 3' end of the antisense strand (Qiagen). SPIO NPs were purchased from OceanNanotech (San Diego, CA). The SPIO NPs had iron oxide cores measuring 15 nm, and outer layers composed of amphiphilic polymer and polyethylenimine groups (PEI). Succinimidyl ester with Alexa Fluor 532 (A532) (Thermo Fisher Scientific) was used to label the SPIO NPs. Clinical-grade doxorubicin and vincristine were provided by the UC Davis Pharmacy. Maphosphamide and clinical-grade cisplatin were purchased from Santa Cruz Biotechnology (Dallas, TX) and the UC Davis Pharmacy, respectively.
siRNA Nanocomplexes
Nanocomplexes were formulated as described previously (10). Briefly, SPIO NPs with PEI groups on the surface were reacted with A532 amine-reactive succinimidyl ester and incubated for 3 h at 4 °C. siRNAs were adsorbed onto the surface of the A532-SPIO NPs by vortexing. The final siRNA nanocomplexes were then resuspended in Opti-mem Reduced Serum Medium (Thermo Fisher Scientific).
The hydrodynamic diameter and zeta potential of the siRNA nanocomplexes were characterized using the Zetasizer Nano ZS (Malvern, UK), as described previously (10). For each measurement, 0.5 mg of SPIO NPs (without A532 labeling) were combined with 0.5 mg of siRNA and resuspended in 1 ml of ultrapure water. A total of three measurements for both size and zeta potential were performed.
Loading efficiency of siRNAs on the SPIO NPs was measured using fluorescent titration curves as described previously (10). After vortexing A488-labeled siRNAs with SPIO NPs (without A532 labeling) in water, the solution was centrifuged at 900g for 4 min. After centrifugation, pelleted nanocomplexes and supernatant were measured for fluorescence with a SpectraMax Paradigm Multi-Mode Microplate Reader (Molecular Devices, Sunnyvale, CA) to determine the amount of siRNA. A total of three experiments were performed.
Apoptosis Assay
Cell apoptosis was measured by annexin V and caspase activity. siRNA nanocomplexes (without A532 labeling) were used. Annexin V was measured by flow cytometry using annexin V conjugated to APC (BD Biosciences, San Diego, CA). Treated cells as described above were stained in their wells with 95 μl of binding buffer and 5 μl of annexin V according to the manufacturer’s instructions. Data were acquired with an FC500 flow cytometer and analyzed with CXP analysis software (Beckman Coulter, San Jose, CA). Caspase activity was measured using the Caspase 3/7 Glo kit (Promega, Madison, WI). Cells were plated in 96-well tissue culture-treated plates with seeding densities of 20,000 cells per 170 μl of growth medium per well. Each sample was mixed with 40 μl of Caspase 3/7 Glo reagent and incubated for 1 h at room temperature according to the manufacturer’s instructions. Samples were analyzed using a Centro LB 960 Microplate Luminometer (Berthold Technologies, Oakridge, TN). These experiments were independently performed three times, in triplicate, for caspase assay.
Cell Viability Following Treatment with siRNA Nanocomplexes and Chemotherapy Drugs
Cells were seeded in complete medium at least 12 h before treatment. The seeding density was 600,000 cells per 2 ml of medium per well or 60,000 cells per 0.5 ml medium per well in 6- or 48-well tissue culture-treated plates, respectively. The siRNA nanocomplexes were added to the cells in Opti-mem, and they were then incubated for 4 h, followed by replacement with fresh medium. Cells were collected for analysis of MXD3 protein expression by immunocytochemistry at the indicated times. This experiment was repeated three times, and for each treatment group and time point wells were prepared in triplicate.
For combination therapy experiments, cells were treated with doxorubicin, vincristine, cisplatin, or maphosphamide at their respective half maximal inhibitory concentration (IC50) for each drug 4 h after siRNA nanocomplex treatment. The IC50 values had been determined from three repeated MTS assays. The average IC50 values for doxorubicin, vincristine, cisplatin, and maphosphamide were 176.93, 33.34, 1,566, and 1,963 ng/ml, respectively, for SK-N-DZ, and 33.82, 4.65, 695, and 804.7 ng/ml, respectively, for SK-N-BE.
Statistical Methods
Statistical analysis was performed using standard linear models implemented in SAS Version 9.4; cell counts were log-transformed. Multiple comparisons of means used SAS PROC MULTTEST, adjusted for multiple comparisons via the Hochberg–Benjamini approach (24). Significant findings in analysis of variance comparisons of means were further examined with Tukey’s studentized range test for experiment-wise control of Type I error. A P value <0.05 was considered significant for all statistical calculations.
Results
MXD3 is Highly Expressed in Neuroblastoma
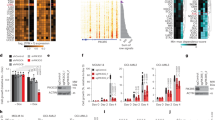
MXD3 expression was evaluated in neuroblastoma cell lines and primary tumor samples based on mRNA and protein levels (Supplementary Figure S1). mRNA levels were measured using RT-qPCR. We used normal human adrenal gland (containing both medulla and cortex) as a control because adrenal gland (medulla) is a common site for neuroblastoma to arise (25). On the basis of our previous study, we also used peripheral blood mononuclear cells (PBMCs) and Jurkat cells, and Reh cells as negative and positive controls, respectively (10). In five neuroblastoma cell lines, MXD3 mRNA expression was substantially and significantly elevated, ranging between 2.6- and 14.6-fold relative to PBMCs or adrenal gland (Supplementary Figure S1a all P<0.001 after Bonferroni adjustment). A public mRNA expression data set of 24 neuroblastoma cell lines (BioGPS, array E-GEOD-28019) also shows a trend of high MXD3 expression relative to PBMCs (http://biogps.org). MXD3 protein expression was also elevated in these cell lines using immunocytochemistry (Supplementary Figure S1b,S1c). Next, MXD3 protein expression was tested in 18 primary human neuroblastoma samples using tumor microarray immunohistochemistry. Normal kidney and liver tissues were used as controls, both of which showed low levels of MXD3 expression. Seven out of 18 samples showed high MXD3 expression (Figure 1). In this group, all seven patients had at least one high-risk factor (age >18 months, advanced stage, N-myc amplification, poorly or undifferentiated histology) (Supplementary Table S1). These results showed that MXD3 is highly expressed in a subset of neuroblastoma at both mRNA and protein levels.
MXD3 is highly expressed in patient neuroblastoma tumor samples. High MXD3 protein expression is observed in a subset (high-risk) of patient tumor samples (images acquired at × 40 magnification) (Supplementary Table S1). Normal kidney and liver tissues were used as controls.
Novel Nanocomplexes are Created Using MXD3 siRNA and SPIO NPs
siRNAs were combined with SPIO NPs based on electrostatic interactions between the NPs and siRNA molecules and characterized by size and charge (Figure 2) (10). The size of the SPIO NPs was 40.4 nm in diameter (polydispersity 0.120). When electrostatically combined with siRNA, the size of the siRNA nanocomplexes was 56.1 nm in diameter (polydispersity 0.399). Surface charges of the SPIO NPs alone and the siRNA nanocomplexes were +58.8 and +42.4 mV, respectively. These results were similar to the previously reported characteristics of NPs (26). High loading efficiency of the siRNA onto the NPs was shown by fluorescence measurements: 95.6% of the siRNA molecules when loaded alone to the NPs, which was an average of 294 molecules of siRNA per SPIO NP, or 79.55 pmol of siRNA per 1 μg of SPIO NP by iron content.
Characterization of siRNA nanocomplexes. Diameter and zeta potential of the siRNA nanocomplexes are shown. The size and zeta potential of the nanocomplexes changed after loading the siRNAs on the superparamagnetic iron oxide (SPIO) nanoparticles (NPs). Top and bottom graphs indicate before and after siRNA loading, respectively.
Intracellular Uptake of siRNA Nanocomplexes Results in MXD3 Knockdown and Apoptosis in Neuroblastoma Cells
We next investigated the in vitro delivery and therapeutic effects of the nanocomplexes in the neuroblastoma cell lines SK-N-DZ and SK-N-BE. These two cell lines were chosen because they showed the highest MXD3 expression among the tested cell lines. After incubating cells for 4 h with the siRNA nanocomplexes, intracellular co-localization of the A488-siRNA and A532-SPIO NPs was observed (Figure 3a), indicating intracellular delivery of the nanocomplexes. In SK-N-DZ, cells treated with the MXD3 siRNA nanocomplexes showed a MXD3 protein knockdown by immunocytochemistry compared with untreated cells (73.4% reduction, P=0.007) or cells treated with the control siRNA nanocomplexes (63% reduction, P=ns) (Figure 3b). MXD3 protein expression was also confirmed using western blotting (Supplementary Figure S2). In SK-N-BE, cells treated with the MXD3 siRNA nanocomplexes also showed a significant MXD3 protein knockdown compared with untreated cells (72% reduction, P⩽0.001) or cells treated with the control siRNA nanocomplexes (68% reduction, P⩽0.001) (Figure 3e).
Intracellular delivery of the MXD3 siRNA nanocomplexes results in MXD3 knockdown and cell growth inhibition in vitro. (a–c) SK-N-DZ and (d–f) SK-N-BE. (a,d) Intracellular co-localization of siRNA (green) and NPs (red) at 4 h after treatment are shown. Untreated cells were shown as control. (b,e) MXD3 protein knockdown is observed in the cells treated with the MXD3 siRNA nanocomplexes at 4 h after treatment. Control siRNA nanocomplex-treated cells showed similar levels of MXD3 expression to untreated cells. (c,f) MXD3 protein knockdown was quantified using mean fluorescence intensity (MFI). Each point represents the average MFI of all measured cells per treatment type, in 4 independent experiments. (c) Untreated vs. MXD3 siRNA nanocomplexes (**) P=0.007. (f) Untreated or control siRNA nanocomplexes vs. MXD3 siRNA nanocomplexes (***) P<0.001.
We then investigated whether the nanocomplex treatment induces apoptosis using annexin V and caspase assays. SK-N-DZ cells treated with MXD3 siRNA nanocomplexes showed a significant increase in the annexin V-positive population when compared with the control groups at both 4 and 8 h after a single treatment (Figure 4a). At 4 h, untreated cells and control siRNA nanocomplex-treated cells were 2.5 and 4.2% positive for annexin V, respectively, whereas MXD3 siRNA nanocomplex-treated cells were 11% positive (P<0.001) (Figure 4b). At 8 h, untreated cells and control siRNA nanocomplex-treated cells were 2.8 and 5.2% positive for annexin V, respectively, whereas MXD3 siRNA nanocomplex-treated cells were 12.3% positive (P<0.001) (Figure 4b). For both time points, there was no significant difference in annexin V between untreated cells and the control siRNA nanocomplex-treated cells. Cells were also analyzed for caspase 3 and 7 activities. Cells treated with MXD3 siRNA nanocomplexes showed a significant increase in the caspase activity compared with the control groups both 4 and 8 h after a single treatment (all P<0.001) (Figure 4c). Of note, there was a significant increase in caspase activity in the control siRNA nanocomplex-treated cells compared with untreated cells at both time points (P<0.001) (Figure 4c). The results in SK-N-BE cells were similar to those in SK-N-DZ cells (Figure 4d). At 4 h, untreated control siRNA nanocomplex-treated cells were 11.2 and 14.5% positive for annexin V, respectively, whereas MXD3 siRNA nanocomplex-treated cells were 22.7% positive. At 8 h, untreated cells and control siRNA nanocomplex-treated cells were 12.1 and 17.1% positive for annexin V, respectively, whereas MXD3 siRNA nanocomplex-treated cells were 35.1% positive (vs. untreated P<0.001, vs. control siRNA nanocomplexes P<0.01) (Figure 4e). For both time points, there was no significant difference between untreated cells and control siRNA nanocomplex-treated cells. Cells treated with MXD3 siRNA nanocomplexes also showed a significant increase in the caspase activity compared with the control groups at both time points (P<0.001) (Figure 4f). Altogether, these results indicate that MXD3 knockdown induced rapid cell apoptosis in SK-N-DZ and SK-N-BE cells.
Treatment with the MXD3 siRNA nanocomplexes increases apoptosis in the SK-N-DZ and SK-N-BE cells. SK-N-DZ (a–c) or SK-N-BE (d–f) cells treated with the MXD3 siRNA nanocomplexes were more annexin V or caspase positive than untreated cells or cells treated with the control siRNA nanocomplexes at both 4 and 8 h after treatment. (a,d) Flow cytometry graphs are shown from one representative experiment out of three independent experiments. (b,e) Quantification of annexin V-positive cells from three experiments. Data as mean±SD. n=3 for each time point. (b) Untreated or control siRNA nanocomplexes vs. MXD3 siRNA nanocomplexes at both time points (***) P<0.001. (e) Untreated vs. MXD3 siRNA nanocomplexes at 8 h (***) P<0.001. Control vs. MXD3 siRNA nanocomplexes at 8 h (**) P<0.01. (c,f) Cell apoptosis measured by caspase 3 and caspase 7 activities are shown. Data include three independent experiments. Data as mean±SD. n=9 for each time point. (c) Untreated vs. control or MXD3 siRNA nanocomplexes, and control vs. MXD3 siRNA nanocomplexes, at both time points (***) P<0.001. (f) Untreated vs. control or MXD3 siRNA nanocomplexes, and control vs. MXD3 siRNA nanocomplexes, at both time points (***) P<0.001.
To confirm that the higher levels of apoptosis we observed were indeed due to MXD3 knockdown and not off-target effects, we tested the treatment on SK-N-DZ cells expressing a rescue construct coding for knockdown-insensitive, wild-type MXD3 (9). SK-N-DZ cells were stably transduced using lentiviral vectors expressing MXD3 shRNA and the rescue sequence (shMXD3Rescue), MXD3 shRNA alone (shMXD3), or a negative control shRNA (shControl) designed to not knock down any known human gene. shMXD3 and shControl sequences were designed to produce the exact siRNA sequences used in the rest of this study. All constructs also included green fluorescent protein (GFP) for analysis of transduction efficiency. Transduction efficiency at 72 h after transduction, measured as GFP expression by flow cytometry, ranged from 67 to 98% (Supplementary Figure S3a). Cells transduced with the shMXD3 showed MXD3 protein knockdown by immunocytochemistry compared with untransduced cells (70.1% reduction, P=0.03) or cells transduced with shControl or shMXD3Rescue (Supplementary Figure S3b,S3c). Cells transduced with shMXD3 also had decreased viable cell counts 72 h after transduction compared with the control groups (Supplementary Figure S3d). Importantly, when treated with MXD3 siRNA nanocomplexes, shMXD3Rescue-transduced cells showed significantly lower caspase activity when compared with untransduced cells or shControl-transduced cells at 4 h (vs. untransduced P=0.0064, vs. shControl P=0.043) and 8 h (vs. untransduced P=0.0024, vs. shControl P<0.001) after a single treatment (Supplementary Figure S3e). These results together indicate that the increase in apoptosis observed in SK-N-DZ cells treated with MXD3 siRNA nanocomplexes was mediated by MXD3 knockdown and not by an off-target effect.
Combination of the MXD3 siRNA Nanocomplexes and Doxorubicin, Vincristine, Cisplatin, or Maphosphamide Shows Additive Cytotoxic Effects
We assessed the therapeutic potential of the MXD3 siRNA nanocomplexes with chemotherapy drugs currently used in neuroblastoma treatment in SK-N-DZ and SK-N-BE in vitro. We added a single dose of doxorubicin, vincristine, cisplatin, or maphosphamide at the respective cell line’s IC50 doses 4 h after treatment with the siRNA nanocomplexes, and measured cytotoxicity after 72 h using a standard 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay.
In SK-N-DZ cells, the average cytotoxicity of the combination therapy (91.2%) was significantly higher than that of nanocomplexes (30.5%) or doxorubicin (61.2%) (both P⩽0.001) (Figure 5a). Similarly, the average cytotoxicity of the combination therapy (52%) was higher than that of nanocomplexes (24.5%, P<0.001) or vincristine (45.5%) (Figure 5b). The cytotoxicity of MXD3 siRNA nanocomplexes, cisplatin, or maphosphamide was an average of 49.2, 44.9, and 41.2%, respectively, whereas the cytotoxicity of the combination of MXD3 siRNA nanocomplexes with either cisplatin or maphosphamide was an average of 70 or 69.3%, respectively (single agents vs. combination all P<0.001) (Figure 5c).
Combination therapy of MXD3 siRNA nanocomplexes with doxorubicin, vincristine, cisplatin, and maphosphamide showed additive cytotoxic effects. SK-N-DZ (a–d) or SK-N-BE (e–h) cells were treated with various drugs as described. (a) Nanocomplexes or doxorubicin only vs. combination (***) P<0.001. (b) Nanocomplexes vs. combination (***) P<0.001. (c) Nanocomplexes or cisplatin only vs. combination (***) P<0.001. (d) Nanocomplexes or maphosphamide vs. combination (***) P<0.001. (e) Nanocomplexes or doxorubicin only vs. combination (***) P<0.001 and (**) P<0.01, respectively. (f) Nanocomplexes or vincristine only vs. combination (***) P<0.001. (g) Nanocomplexes or cisplatin only vs. combination (***) P<0.001. (h) Nanocomplexes or maphosphamide only vs. combination (***) P<0.001.
In SK-N-BE, the average cytotoxicity of the combination therapy (83%) was significantly higher than that of nanocomplexes (32.6%, P<0.001) or doxorubicin (60%, P<0.01) (Figure 5e). The average cytotoxicity of the combination therapy (63.7%) was significantly higher than that of nanocomplexes (28.5%) or vincristine (46.2%) (both P<0.001) (Figure 5f). The cytotoxicity of MXD3 siRNA nanocomplexes, cisplatin, or maphosphamide was an average of 42.3, 50, and 53%, respectively, whereas the cytotoxicity of the combination of MXD3 siRNA nanocomplexes with either cisplatin or maphosphamide was an average of 74.5 or 72.9%, respectively (single agents vs. combination, all P<0.001) (Figure 5g). These results demonstrate the potential of using the MXD3 siRNA nanocomplexes as part of current chemotherapy regimens for neuroblastoma treatment.
Discussion
Neuroblastoma is a heterogeneous disease, ranging from a type that spontaneously regresses to high-risk types, which require intensive treatment (2). Over the past several decades, greater understanding of the biology of neuroblastoma and the development of treatments based on risk type has improved the overall prognosis. However, patients with high-risk disease continue to have a very poor prognosis despite the most intensive treatments currently available (27). In addition, these treatments have significant short- and long-term adverse effects (28). The development of targeted therapies can be one alternative to increase current treatment efficacy without adverse effects. Although there are few targeted therapies currently used for neuroblastoma, recent introduction of an anti-GD2 antibody dinutuximab, targeting GD2 receptors on the tumor cell surface and inducing immune-mediated cytotoxicity, has shown promising results (29). Several drugs targeting genes are also currently being evaluated in clinical trials. The anti-protozoan drug difluromethylornithine, for example, has been recently investigated in clinical trials as an inhibitor of polyamine synthesis in neuroblastoma with MYC-dependent proliferation (30). Crizotinib, an ALK-inhibitor, is currently undergoing Phase I/II trials as a treatment for solid refractory tumors, including neuroblastoma (31).
To further improve the outcome of high-risk neuroblastoma, it is necessary to use combination therapies of drugs with different targets. Our study discovered a potential new molecular target, MXD3. We demonstrated high MXD3 expression in both cell lines and primary tumor samples at either mRNA or protein levels (Supplementary Figure S1 and Figure 1). Interestingly, primary tumor samples taken from 18 patients with neuroblastoma showed a trend between elevated MXD3 expression and high-risk status: all 7 patient samples with high MXD3 expression had more than one high-risk factor (Figure 1 and Supplementary Table 1). Publicly available microarray data on patients with neuroblastoma also indicate that MXD3 expression is associated with poor outcomes (R2: Genomics Analysis and Visualization Platform). Our study and those of others showed that MXD3 expression is very limited in normal tissues, including adrenal glands and hematopoietic stem cells (www.genecards.org) (10), which makes MXD3 an ideal therapeutic target. We demonstrated that inhibition of MXD3 induced cell apoptosis in neuroblastoma cells (Figure 4) and that the cell apoptosis was indeed mediated by MXD3 knockdown with the siRNA (Supplementary Figure S3).
We have developed a successful compound to deliver siRNA into tumor cells using SPIO NPs. Our conjugate showed 95.6% loading efficiency of siRNA (an average of 294 molecules of siRNA per SPIO NP) (10). One study showed that siRNA was conjugated to SPIO NPs, using a succinimidyl 3-(2-pyridyldithio) propionate (SPDP) linker and thiol-modified siRNA, in ratios of 4.2 pmol of siRNA per 1 μg of SPIO NP by iron content (22). Our siRNA nanocomplexes had a ratio of about 77 pmol of siRNA per 1 μg of SPIO NP, by iron content. The difference in the amount of siRNA on NPs between ours and this study could be because of conjugation efficiency, as the siRNA nanocomplexes used in our study were made using electrostatic interactions to adsorb siRNA onto SPIO NPs. In addition to our nanocomplex’s high loading efficiency, the siRNA nanocomplexes were able to enter intact (co-localization of fluorescent markers) into the cells in vitro (Figure 3a). Last, cells treated with the MXD3 siRNA nanocomplexes showed knockdown of MXD3 expression compared with untreated and control siRNA nanocomplexes (Figure 3b). These results together suggest that the nanocomplexes were taken up and released functional siRNA intracellularly. For this delivery platform to work successfully in vivo, we expect that protective modifications to the siRNA (32) or to the surface of the nanocomplexes (33) will be necessary.
MXD3 knockdown in neuroblastoma cells led to a significant increase in apoptosis measured by annexin V and caspase activity (Figure 4). The control siRNA nanocomplexes showed some increase in apoptosis compared with untreated cells, which is likely due to the properties of the SPIO NPs. We expect that with further modifications to the NP surface nonspecific toxicity of the nanocomplexes will decrease. For instance, SPIO NPs with poly(ɛ-caprolactone)-poly(ethylene glycol) (PCL-PEG)-modified surfaces for increased biocompatibility can be used to deliver cancer drugs such as methotrexate in vitro (34). Another study conducted with doxorubicin-loaded SPIO NPs used a bovine serum albumin shell to encapsulate doxorubicin, along with a PEG-modified outer shell (35). Additional modifications include dextran and aminosilane-modified outer groups, which have been shown to reduce the cytotoxicity of SPIOs with amine groups (36).
We demonstrated that the MXD3 siRNA nanocomplexes are not only therapeutic as a single agent but also have the potential to be combined with currently used chemotherapy drugs. For both SK-N-DZ and SK-N-BE, the cytotoxicity of all four drugs, doxorubicin, vincristine, cisplatin, or maphosphamide, was increased when combined with MXD3 siRNA nanocomplexes (Figure 5). These results are highly encouraging, as we only investigated single treatments of each drug, at its IC50 dose, with MXD3 siRNA nanocomplexes. It is possible to further increase cytotoxicity by optimizing dose and schedule of these combination therapies, combining multiple drugs or including other drugs. It is also possible that the MXD3 siRNA nanocomplexes may replace some of the current chemotherapy drugs and minimize toxic adverse effects. DNA-intercalating chemotherapy drugs, such as doxorubicin, and alkylators, such as cisplatin, have been reported to work synergistically with deacetylase inhibitors, which induce apoptosis in cells (37). Further studies are necessary with more samples and different regimens.
In conclusion, we have shown that MXD3 is a new potential molecular target for treating neuroblastoma, particularly for high-risk type, which has high MXD3 expression, and demonstrated a proof-of-concept for a drug that targets MXD3 using siRNA-mediated knockdown. Future studies will focus on introducing siRNA structure modification to protect them from endonucleases (32), optimizing SPIO NP surface chemistries to minimize its inherent toxicity (36, 38), and using a targeted molecule unique to neuroblastoma to deliver the nanocomplexes more specifically to the tumor cells (39).
References
Schleiermacher G, Janoueix-Lerosey I, Delattre O . Recent insights into the biology of neuroblastoma. Int J Cancer 2014;135:2249–2261.
Neuroblastoma Treatment (PDQ(R)). Health Professional Version. PDQ Cancer Information Summaries: Bethesda, MD, 2002.
Cohen LE, Gordon JH, Popovsky EY et al, Late effects in children treated with intensive multimodal therapy for high-risk neuroblastoma: high incidence of endocrine and growth problems. Bone Marrow Transplant 2014;49:502–508.
Young SW, Stenzel M, Jia-Lin Y . Nanoparticle-siRNA: a potential cancer therapy? Crit Rev Oncol Hematol 2016;98:159–169.
Koldehoff M, Steckel NK, Beelen DW, Elmaagacli AH . Therapeutic application of small interfering RNA directed against bcr-abl transcripts to a patient with imatinib-resistant chronic myeloid leukaemia. Clin Exp Med 2007;7:47–55.
Golan T, Khvalevsky EZ, Hubert A et al, RNAi therapy targeting KRAS in combination with chemotherapy for locally advanced pancreatic cancer patients. Oncotarget 2015;6:24560–24570.
Di Paolo D, Yang D, Pastorino F et al, New therapeutic strategies in neuroblastoma: combined targeting of a novel tyrosine kinase inhibitor and liposomal siRNAs against ALK. Oncotarget 2015;6:28774–28789.
Evans L, Chen L, Milazzo G et al, SKP2 is a direct transcriptional target of MYCN and a potential therapeutic target in neuroblastoma. Cancer Lett 2015;363:37–45.
Barisone GA, Satake N, Lewis C et al, Loss of MXD3 induces apoptosis of Reh human precursor B acute lymphoblastic leukemia cells. Blood Cells Mol Dis 2015;54:329–335.
Satake N, Duong C, Chen C et al, Targeted therapy with MXD3 siRNA, anti-CD22 antibody and nanoparticles for precursor B-cell acute lymphoblastic leukaemia. Br J Haematol 2014;167:487–499.
Satake N, Duong C, Yoshida S et al, Novel targeted therapy for precursor B cell acute lymphoblastic leukemia: anti-CD22 antibody-MXD3 antisense oligonucleotide conjugate. Mol Med 2016;22:632–642.
Fox EJ, Wright SC . The transcriptional repressor gene Mad3 is a novel target for regulation by E2F1. Biochem J 2003;370:307–313.
Barisone GA, Yun JS, Diaz E . From cerebellar proliferation to tumorigenesis: new insights into the role of Mad3. Cell Cycle 2008;7:423–427.
Kim HJ, Kim A, Miyata K, Kataoka K . Recent progress in development of siRNA delivery vehicles for cancer therapy. Adv Drug Deliv Rev 2016;104:61–77.
Whitehead KA, Langer R, Anderson DG . Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov 2009;8:129–138.
Nazir S, Hussain T, Ayub A, Rashid U, MacRobert AJ . Nanomaterials in combating cancer: therapeutic applications and developments. Nanomedicine 2014;10:19–34.
Schultheis B, Strumberg D, Santel A et al, First-in-human phase I study of the liposomal RNA interference therapeutic Atu027 in patients with advanced solid tumors. J Clin Oncol 2014;32:4141–4148.
Zuckerman JE, Gritli I, Tolcher A et al, Correlating animal and human phase Ia/Ib clinical data with CALAA-01, a targeted, polymer-based nanoparticle containing siRNA. Proc Natl Acad Sci USA 2014;111:11449–11454.
Wicki A, Witzigmann D, Balasubramanian V, Huwyler J . Nanomedicine in cancer therapy: challenges, opportunities, and clinical applications. J Control Release 2015;200:138–157.
Ittrich H, Peldschus K, Raabe N, Kaul M, Adam G . Superparamagnetic iron oxide nanoparticles in biomedicine: applications and developments in diagnostics and therapy. Rofo 2013;185:1149–1166.
Chen B, Wu W, Wang X . Magnetic iron oxide nanoparticles for tumor-targeted therapy. Curr Cancer Drug Targets 2011;11:184–189.
Wang P, Yigit MV, Medarova Z et al, Combined small interfering RNA therapy and in vivo magnetic resonance imaging in islet transplantation. Diabetes 2011;60:565–571.
Burgess A, Vigneron S, Brioudes E, Labbe JC, Lorca T, Castro A . Loss of human Greatwall results in G2 arrest and multiple mitotic defects due to deregulation of the cyclin B-Cdc2/PP2A balance. Proc Natl Acad Sci USA 2010;107:12564–12569.
Hochberg Y, Benjamini Y . More powerful procedures for multiple significance testing. Stat Med 1990;9:811–818.
Herrera JM, Krebs A, Harris P, Barriga F . Childhood tumors. Surg Clin North Am 2000;80:747–760 xii.
Uchegbu IF, Siew A . Nanomedicines and nanodiagnostics come of age. J Pharm Sci 2013;102:305–310.
Yalcin B, Kremer LC, van Dalen EC . High-dose chemotherapy and autologous haematopoietic stem cell rescue for children with high-risk neuroblastoma. Cochrane Database Syst Rev 2015;10:CD006301.
Choi DK, Schmidt ML . Chemotherapy in children with head and neck cancers: perspectives and review of current therapies. Oral Maxillofac Surg Clin North Am 2016;28:127–138.
Horta ZP, Goldberg JL, Sondel PM . Anti-GD2 mAbs and next-generation mAb-based agents for cancer therapy. Immunotherapy 2016;8:1097–1117.
Bassiri H, Benavides A, Haber M, Gilmour SK, Norris MD, Hogarty MD . Translational development of difluoromethylornithine (DFMO) for the treatment of neuroblastoma. Transl Pediatr 2015;4:226–238.
Mosse YP, Lim MS, Voss SD et al, Safety and activity of crizotinib for paediatric patients with refractory solid tumours or anaplastic large-cell lymphoma: a Children's Oncology Group phase 1 consortium study. Lancet Oncol 2013;14:472–480.
Ku SH, Jo SD, Lee YK, Kim K, Kim SH . Chemical and structural modifications of RNAi therapeutics. Adv Drug Deliv Rev 2016;104:16–28.
Khodabandehloo H, Zahednasab H, Ashrafi Hafez A . Nanocarriers usage for drug delivery in cancer therapy. Iran J Cancer Prev 2016;9:e3966.
Jahangiri S, Akbarzadeh A . Preparation and in vitro evaluation of Methotrexate-loaded magnetic nanoparticles modified with biocompatible copolymers. Artif Cells Nanomed Biotechnol 2016;44:1733–1740.
Semkina A, Abakumov M, Grinenko N et al, Core-shell-corona doxorubicin-loaded superparamagnetic Fe3O4 nanoparticles for cancer theranostics. Colloids Surf B Biointerfaces 2015;136:1073–1080.
Wang YX, Xuan S, Port M, Idee JM . Recent advances in superparamagnetic iron oxide nanoparticles for cellular imaging and targeted therapy research. Curr Pharm Des 2013;19:6575–6593.
Stiborova M, Eckschlager T, Poljakova J et al, The synergistic effects of DNA-targeted chemotherapeutics and histone deacetylase inhibitors as therapeutic strategies for cancer treatment. Curr Med Chem 2012;19:4218–4238.
Gupta AK, Wells S . Surface-modified superparamagnetic nanoparticles for drug delivery: preparation, characterization, and cytotoxicity studies. IEEE Trans Nanobiosci 2004;3:66–73.
Berois N, Osinaga E . Glycobiology of neuroblastoma: impact on tumor behavior, prognosis, and therapeutic strategies. Front Oncol 2014;4:114.
Acknowledgements
We thank Wassim N. Hmeidan and Amanda R. Favila for technical assistance on the experiments and formulating figures and a table.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
STATEMENT OF FINANCIAL SUPPORT
This work was supported by research funding from the Keaton Raphael Memorial Foundation, The Hartwell Foundation, Hyundai Hope on Wheels, National Center for Advancing Translational Sciences, NIH, through Grant #UL1 TR000002, and CTSC-MCRTP (Satake) and the California Institute for Regenerative Medicine, and NIH Transformative Grant R01GM099688 (Nolta). Statistical support was provided through the Biostatistics Shared Resource, UC Davis Comprehensive Cancer Center Support Grant P30CA093373-04.
Supplementary material is linked to the online version of the paper at
Rights and permissions
About this article
Cite this article
Duong, C., Yoshida, S., Chen, C. et al. Novel targeted therapy for neuroblastoma: silencing the MXD3 gene using siRNA. Pediatr Res 82, 527–535 (2017). https://doi.org/10.1038/pr.2017.74
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/pr.2017.74