Abstract
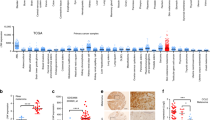
We provide evidence that the IFN-regulated member of the Schlafen (SLFN) family of proteins, SLFN5, promotes the malignant phenotype in glioblastoma multiforme (GBM). Our studies indicate that SLFN5 expression promotes motility and invasiveness of GBM cells, and that high levels of SLFN5 expression correlate with high-grade gliomas and shorter overall survival in patients suffering from GBM. In efforts to uncover the mechanism by which SLFN5 promotes GBM tumorigenesis, we found that this protein is a transcriptional co-repressor of STAT1. Type-I IFN treatment triggers the interaction of STAT1 with SLFN5, and the resulting complex negatively controls STAT1-mediated gene transcription via interferon stimulated response elements. Thus, SLFN5 is both an IFN-stimulated response gene and a repressor of IFN-gene transcription, suggesting the existence of a negative-feedback regulatory loop that may account for suppression of antitumor immune responses in glioblastoma.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev 2007; 21: 2683–2710.
Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006; 444: 756–760.
Desjardins A, Rich JN, Quinn JA, Vredenburgh J, Gururangan S, Sathornsumetee S et al. Chemotherapy and novel therapeutic approaches in malignant glioma. Front Biosci 2005; 10: 2645–2668.
Platanias LC . Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol 2005; 5: 375–386.
Platanias LC . Interferons and their antitumor properties. J Interferon Cytokine Res 2013; 33: 143–144.
Wakabayashi T, Hatano N, Kajita Y, Yoshida T, Mizuno M, Taniguchi K et al. Initial and maintenance combination treatment with interferon-beta, MCNU (Ranimustine), and radiotherapy for patients with previously untreated malignant glioma. J Neurooncol 2000; 49: 57–62.
Natsume A, Ishii D, Wakabayashi T, Tsuno T, Hatano H, Mizuno M et al. IFN-beta down-regulates the expression of DNA repair gene MGMT and sensitizes resistant glioma cells to temozolomide. Cancer Res 2005; 65: 7573–7579.
Motomura K, Natsume A, Kishida Y, Higashi H, Kondo Y, Nakasu Y et al. Benefits of interferon-beta and temozolomide combination therapy for newly diagnosed primary glioblastoma with the unmethylated MGMT promoter: a multicenter study. Cancer 2011; 117: 1721–1730.
Bradley NJ, Darling JL, Oktar N, Bloom HJ, Thomas DG, Davies AJ . The failure of human leukocyte interferon to influence the growth of human glioma cell populations: in vitro and in vivo studies. Br J Cancer 1983; 48: 819–825.
Fish EN, Platanias LC . Interferon receptor signaling in malignancy: a network of cellular pathways defining biological outcomes. Mol Cancer Res 2014; 12: 1691–1703.
Porritt RA, Hertzog PJ . Dynamic control of type I IFN signalling by an integrated network of negative regulators. Trends Immunol 2015; 36: 150–160.
Mavrommatis E, Fish EN, Platanias LC . The schlafen family of proteins and their regulation by interferons. J Interferon Cytokine Res 2013; 33: 206–210.
Katsoulidis E, Mavrommatis E, Woodard J, Shields MA, Sassano A, Carayol N et al. Role of interferon {alpha} (IFN{alpha})-inducible Schlafen-5 in regulation of anchorage-independent growth and invasion of malignant melanoma cells. J Biol Chem 2010; 285: 40333–40341.
Sassano A, Mavrommatis E, Arslan AD, Kroczynska B, Beauchamp EM, Khuon S et al. Human Schlafen 5 (SLFN5) is a regulator of motility and invasiveness of renal cell carcinoma cells. Mol Cell Biol 2015; 35: 2684–2698.
Geserick P, Kaiser F, Klemm U, Kaufmann SH, Zerrahn J . Modulation of T cell development and activation by novel members of the Schlafen (slfn) gene family harbouring an RNA helicase-like motif. Int Immunol 2004; 16: 1535–1548.
Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007; 9: 166–180.
Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell 2006; 9: 287–300.
Madhavan S, Zenklusen JC, Kotliarov Y, Sahni H, Fine HA, Buetow K . Rembrandt: helping personalized medicine become a reality through integrative translational research. Mol Cancer Res 2009; 7: 157–167.
Srikanth M, Das S, Berns EJ, Kim J, Stupp SI, Kessler JA . Nanofiber-mediated inhibition of focal adhesion kinase sensitizes glioma stemlike cells to epidermal growth factor receptor inhibition. Neuro Oncol 2013; 15: 319–329.
Feng H, Lopez GY, Kim CK, Alvarez A, Duncan CG, Nishikawa R et al. EGFR phosphorylation of DCBLD2 recruits TRAF6 and stimulates AKT-promoted tumorigenesis. J Clin Invest 2014; 124: 3741–3756.
Vinci M, Gowan S, Boxall F, Patterson L, Zimmermann M, Court W et al. Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol 2012; 10: 29.
Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science 2008; 321: 956–960.
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010; 28: 511–515.
Zhang Z, Pal S, Bi Y, Tchou J, Davuluri RV . Isoform level expression profiles provide better cancer signatures than gene level expression profiles. Genome Med 2013; 5: 33.
Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L . Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 2013; 31: 46–53.
Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H et al. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res 2013; 41: D1040–D1046.
Wang Z, Cao CJ, Huang LL, Ke ZF, Luo CJ, Lin ZW et al. EFEMP1 promotes the migration and invasion of osteosarcoma via MMP-2 with induction by AEG-1 via NF-kappaB signaling pathway. Oncotarget 2015; 6: 14191–14208.
Ji F, Wang Y, Qiu L, Li S, Zhu J, Liang Z et al. Hypoxia inducible factor 1alpha-mediated LOX expression correlates with migration and invasion in epithelial ovarian cancer. Int J Oncol 2013; 42: 1578–1588.
Nagato S, Nakagawa K, Harada H, Kohno S, Fujiwara H, Sekiguchi K et al. Downregulation of laminin alpha4 chain expression inhibits glioma invasion in vitro and in vivo. Int J Cancer 2005; 117: 41–50.
Levy DE, Kessler DS, Pine R, Reich N, Darnell JE Jr . Interferon-induced nuclear factors that bind a shared promoter element correlate with positive and negative transcriptional control. Genes Dev 1988; 2: 383–393.
Reich N, Evans B, Levy D, Fahey D, Knight E Jr, Darnell JE Jr . Interferon-induced transcription of a gene encoding a 15-kDa protein depends on an upstream enhancer element. Proc Natl Acad Sci USA 1987; 84: 6394–6398.
Kramer A, Green J, Pollard J Jr, Tugendreich S . Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014; 30: 523–530.
Parker BS, Rautela J, Hertzog PJ . Antitumour actions of interferons: implications for cancer therapy. Nat Rev Cancer 2016; 16: 131–144.
Ritchie KJ, Zhang DE . ISG15: the immunological kin of ubiquitin. Semin Cell Dev Biol 2004; 15: 237–246.
Gil MP, Bohn E, O'Guin AK, Ramana CV, Levine B, Stark GR et al. Biologic consequences of Stat1-independent IFN signaling. Proc Natl Acad Sci USA 2001; 98: 6680–6685.
Shuai K, Liu B . Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol 2003; 3: 900–911.
Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res 2004; 64: 7011–7021.
Eckerdt F, Alvarez A, Bell J, Arvanitis C, Iqbal A, Arslan AD et al. A simple, low-cost staining method for rapid-throughput analysis of tumor spheroids. Biotechniques 2016; 60: 43–46.
Yoon CH, Kim MJ, Kim RK, Lim EJ, Choi KS, An S et al. c-Jun N-terminal kinase has a pivotal role in the maintenance of self-renewal and tumorigenicity in glioma stem-like cells. Oncogene 2012; 31: 4655–4666.
Zhu T, Li X, Luo L, Wang X, Li Z, Xie P et al. Reversion of malignant phenotypes of human glioblastoma cells by beta-elemene through beta-catenin-mediated regulation of stemness-, differentiation- and epithelial-to-mesenchymal transition-related molecules. J Transl Med 2015; 13: 356.
Bustos O, Naik S, Ayers G, Casola C, Perez-Lamigueiro MA, Chippindale PT et al. Evolution of the Schlafen genes, a gene family associated with embryonic lethality, meiotic drive, immune processes and orthopoxvirus virulence. Gene 2009; 447: 1–11.
Neumann B, Zhao L, Murphy K, Gonda TJ . Subcellular localization of the Schlafen protein family. Biochem Biophys Res Commun 2008; 370: 62–66.
Li M, Kao E, Gao X, Sandig H, Limmer K, Pavon-Eternod M et al. Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature 2012; 491: 125–128.
Zoppoli G, Regairaz M, Leo E, Reinhold WC, Varma S, Ballestrero A et al. Putative DNA/RNA helicase Schlafen-11 (SLFN11) sensitizes cancer cells to DNA-damaging agents. Proc Natl Acad Sci USA 2012; 109: 15030–15035.
Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012; 483: 603–607.
Companioni Napoles O, Tsao AC, Sanz-Anquela JM, Sala N, Bonet C, Pardo ML et al. SCHLAFEN 5 expression correlates with intestinal metaplasia that progresses to gastric cancer. J Gastroenterol 2016; 52: 39–49.
Acknowledgements
The work was supported in part by NIH grants CA161196, CA77816 and CA155566 and by grant 5I01CX000916 from the Department of Veterans Affairs. ADA was supported in part by NIH training grant T32CA070085, DS was supported in part by NIH training grant T32CA080621, and PL was supported in part by National Science Centre, Poland Grant 2016/22/M/NZ2/00548. We thank Dr John A. Kessler for providing the JK18 and JK46 GSC lines.
Data and materials availability
The microarray data was deposited to the Gene Expression Omnibus (GEO) repository under accession number GSE88771. The RNA-Seq data was deposited to the NCBI's Sequence Read Archive (SRA) repository under the registered BioProject PRJNA341338. The authors declare that all data supporting the findings of this study are available within the article and its Supplementary Information files are available from the corresponding author upon request.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Arslan, A., Sassano, A., Saleiro, D. et al. Human SLFN5 is a transcriptional co-repressor of STAT1-mediated interferon responses and promotes the malignant phenotype in glioblastoma. Oncogene 36, 6006–6019 (2017). https://doi.org/10.1038/onc.2017.205
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.205
This article is cited by
-
SLFN5 promotes reversible epithelial and mesenchymal transformation in ovarian cancer
Journal of Ovarian Research (2023)
-
Structural, molecular, and functional insights into Schlafen proteins
Experimental & Molecular Medicine (2022)
-
Schlafen 5 as a novel therapeutic target in pancreatic ductal adenocarcinoma
Oncogene (2021)
-
Comparative proteomics identifies Schlafen 5 (SLFN5) as a herpes simplex virus restriction factor that suppresses viral transcription
Nature Microbiology (2021)
-
A multi-omics analysis reveals the unfolded protein response regulon and stress-induced resistance to folate-based antimetabolites
Nature Communications (2020)