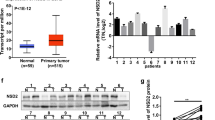
Abstract
Lysine-specific demethylase 1 (LSD1), which has been considered as a potential therapeutic target in human cancer, has been known to regulate many biological functions through its non-histone substrates. Although LSD1-induced hypoxia-inducible factor alpha (HIF1α) demethylation has recently been proposed, the effect of LSD1 on the relationship between HIF1α post-translational modifications (PTMs) and HIF1α-induced tumor angiogenesis remains to be elucidated. Here, we identify a new methylation site of the HIF1α protein antagonized by LSD1 and the interplay between HIF1α protein methylation and other PTMs in regulating tumor angiogenesis. LSD1 demethylates HIF1α at lysine (K) 391, which protects HIF1α against ubiquitin-mediated protein degradation. LSD1 also directly suppresses PHD2-induced HIF1α hydroxylation, which has a mutually dependent interplay with Set9-mediated HIF1α methylation. Moreover, the HIF1α acetylation that occurs in a HIF1α methylation-dependent manner is inhibited by the LSD1/NuRD complex. HIF1α stabilized by LSD1 cooperates with CBP and MTA1 to enhance vascular endothelial growth factor (VEGF)-induced tumor angiogenesis. Thus, LSD1 is a key regulator of HIF1α/VEGF-mediated tumor angiogenesis by antagonizing the crosstalk between PTMs involving HIF1α protein degradation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
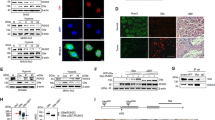
References
Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004; 119: 941–953.
Lee MG, Wynder C, Cooch N, Shiekhattar R . An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 2005; 437: 432–435.
Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y . Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell 2005; 19: 857–864.
Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell 2009; 138: 660–672.
Cho HS, Suzuki T, Dohmae N, Hayami S, Unoki M, Yoshimatsu M et al. Demethylation of RB regulator MYPT1 by histone demethylase LSD1 promotes cell cycle progression in cancer cells. Cancer Res 2011; 71: 655–660.
Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet 2009; 41: 125–129.
Kontaki H, Talianidis I . Lysine methylation regulates E2F1-induced cell death. Mol Cell 2010; 39: 152–160.
Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M et al. p53 is regulated by the lysine demethylase LSD1. Nature 2007; 449: 105–108.
Schulte JH, Lim S, Schramm A, Friedrichs N, Koster J, Versteeg R et al. Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy. Cancer Res 2009; 69: 2065–2071.
Kahl P, Gullotti L, Heukamp LC, Wolf S, Friedrichs N, Vorreuther R et al. Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence. Cancer Res 2006; 66: 11341–11347.
Lim S, Janzer A, Becker A, Zimmer A, Schule R, Buettner R et al. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis 2010; 31: 512–520.
Shi Y . Histone lysine demethylases: emerging roles in development, physiology and disease. Nat Rev Genet 2007; 8: 829–833.
Lin T, Ponn A, Hu X, Law BK, Lu J . Requirement of the histone demethylase LSD1 in Snai1-mediated transcriptional repression during epithelial-mesenchymal transition. Oncogene 2010; 29: 4896–4904.
Li Q, Shi L, Gui B, Yu W, Wang J, Zhang D et al. Binding of the JmjC demethylase JARID1B to LSD1/NuRD suppresses angiogenesis and metastasis in breast cancer cells by repressing chemokine CCL14. Cancer Res 2011; 71: 6899–6908.
Ferrari-Amorotti G, Fragliasso V, Esteki R, Prudente Z, Soliera AR, Cattelani S et al. Inhibiting interactions of lysine demethylase LSD1 with Snail/Slug blocks cancer cell invasion. Cancer Res 2012; 73: 235–245.
Kashyap V, Ahmad S, Nilsson EM, Helczynski L, Kenna S, Persson JL et al. The lysine specific demethylase-1 (LSD1/KDM1A) regulates VEGF-A expression in prostate cancer. Mol Oncol 2013; 7: 555–566.
Qin Y, Zhu W, Xu W, Zhang B, Shi S, Ji S et al. LSD1 sustains pancreatic cancer growth via maintaining HIF1alpha-dependent glycolytic process. Cancer Lett 2014; 347: 225–232.
Arany Z, Huang LE, Eckner R, Bhattacharya S, Jiang C, Goldberg MA et al. An essential role for p300/CBP in the cellular response to hypoxia. Proc Natl Acad Sci USA 1996; 93: 12969–12973.
Rankin EB, Giaccia AJ . The role of hypoxia-inducible factors in tumorigenesis. Cell Death Differ 2008; 15: 678–685.
Jeong JW, Bae MK, Ahn MY, Kim SH, Sohn TK, Bae MH et al. Regulation and destabilization of HIF-1alpha by ARD1-mediated acetylation. Cell 2002; 111: 709–720.
Cheng J, Kang X, Zhang S, Yeh ET . SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell 2007; 131: 584–595.
Liu X, Chen Z, Xu C, Leng X, Cao H, Ouyang G et al. Repression of hypoxia-inducible factor alpha signaling by Set7-mediated methylation. Nucleic Acids Res 2015; 43: 5081–5098.
Kim Y, Nam HJ, Lee J, Park do Y, Kim C, Yu YS et al. Methylation-dependent regulation of HIF-1alpha stability restricts retinal and tumour angiogenesis. Nat Commun 2016; 7: 10347.
Gaughan L, Stockley J, Wang N, McCracken SR, Treumann A, Armstrong K et al. Regulation of the androgen receptor by SET9-mediated methylation. Nucleic Acids Res 2011; 39: 1266–1279.
Ko S, Ahn J, Song CS, Kim S, Knapczyk-Stwora K, Chatterjee B . Lysine methylation and functional modulation of androgen receptor by Set9 methyltransferase. Mol Endocrinol 2011; 25: 433–444.
Page EL, Chan DA, Giaccia AJ, Levine M, Richard DE . Hypoxia-inducible factor-1alpha stabilization in nonhypoxic conditions: role of oxidation and intracellular ascorbate depletion. Mol Biol Cell 2008; 19: 86–94.
Yoo YG, Kong G, Lee MO . Metastasis-associated protein 1 enhances stability of hypoxia-inducible factor-1alpha protein by recruiting histone deacetylase 1. EMBO J 2006; 25: 1231–1241.
Zhang XY, DeSalle LM, Patel JH, Capobianco AJ, Yu D, Thomas-Tikhonenko A et al. Metastasis-associated protein 1 (MTA1) is an essential downstream effector of the c-MYC oncoprotein. Proc Natl Acad Sci USA 2005; 102: 13968–13973.
Amente S, Bertoni A, Morano A, Lania L, Avvedimento EV, Majello B . LSD1-mediated demethylation of histone H3 lysine 4 triggers Myc-induced transcription. Oncogene 2010; 29: 3691–3702.
Nair SS, Li DQ, Kumar R . A core chromatin remodeling factor instructs global chromatin signaling through multivalent reading of nucleosome codes. Mol Cell 2013; 49: 704–718.
Black JC, Van Rechem C, Whetstine JR . Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell 2012; 48: 491–507.
Yang J, Huang J, Dasgupta M, Sears N, Miyagi M, Wang B et al. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc Natl Acad Sci USA 2010; 107: 21499–21504.
Zhang X, Wen H, Shi X . Lysine methylation: beyond histones. Acta Biochim Biophys Sin 2012; 44: 14–27.
Hino S, Kohrogi K, Nakao M . Histone demethylase LSD1 controls the phenotypic plasticity of cancer cells. Cancer Sci 2016; 107: 1187–1192.
Morera L, Lubbert M, Jung M . Targeting histone methyltransferases and demethylases in clinical trials for cancer therapy. Clin Epigenetics 2016; 8: 57.
Serce N, Gnatzy A, Steiner S, Lorenzen H, Kirfel J, Buettner R . Elevated expression of LSD1 (Lysine-specific demethylase 1) during tumour progression from pre-invasive to invasive ductal carcinoma of the breast. BMC Clin Pathol 2012; 12: 13.
Nicolson GL, Nawa A, Toh Y, Taniguchi S, Nishimori K, Moustafa A . Tumor metastasis-associated human MTA1 gene and its MTA1 protein product: role in epithelial cancer cell invasion, proliferation and nuclear regulation. Clin Exp Metastasis 2003; 20: 19–24.
Pakala SB, Singh K, Reddy SD, Ohshiro K, Li DQ, Mishra L et al. TGF-beta1 signaling targets metastasis-associated protein 1, a new effector in epithelial cells. Oncogene 2011; 30: 2230–2241.
Park JH, Lee JY, Shin DH, Jang KS, Kim HJ, Kong G . Loss of Mel-18 induces tumor angiogenesis through enhancing the activity and expression of HIF-1alpha mediated by the PTEN/PI3K/Akt pathway. Oncogene 2011; 30: 4578–4589.
Cho MH, Park JH, Choi HJ, Park MK, Won HY, Park YJ et al. DOT1L cooperates with the c-Myc-p300 complex to epigenetically derepress CDH1 transcription factors in breast cancer progression. Nat Commun 2015; 6: 7821.
Kouskouti A, Scheer E, Staub A, Tora L, Talianidis I . Gene-specific modulation of TAF10 function by SET9-mediated methylation. Mol Cell 2004; 14: 175–182.
Lee MH, Na H, Kim EJ, Lee HW, Lee MO . Poly(ADP-ribosyl)ation of p53 induces gene-specific transcriptional repression of MTA1. Oncogene 2012; 31: 5099–5107.
Acknowledgements
This work was supported by National Research Foundation of Korea (NRF) grants funded by the Korean government (No. 2015R1A2A1A10052578).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Lee, JY., Park, JH., Choi, HJ. et al. LSD1 demethylates HIF1α to inhibit hydroxylation and ubiquitin-mediated degradation in tumor angiogenesis. Oncogene 36, 5512–5521 (2017). https://doi.org/10.1038/onc.2017.158
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.158
This article is cited by
-
Lysine demethylase LSD1 is associated with stemness in EBV-positive B cell lymphoma
Scientific Reports (2024)
-
Astragaloside IV restrains pyroptosis and fibrotic development of pulmonary artery smooth muscle cells to ameliorate pulmonary artery hypertension through the PHD2/HIF1α signaling pathway
BMC Pulmonary Medicine (2023)
-
Role of NAD+ and FAD in Ischemic Stroke Pathophysiology: An Epigenetic Nexus and Expanding Therapeutic Repertoire
Cellular and Molecular Neurobiology (2023)
-
Histone methyltransferase SETD2 inhibits M1 macrophage polarization and glycolysis by suppressing HIF-1α in sepsis-induced acute lung injury
Medical Microbiology and Immunology (2023)
-
The assembly of mammalian SWI/SNF chromatin remodeling complexes is regulated by lysine-methylation dependent proteolysis
Nature Communications (2022)