Abstract
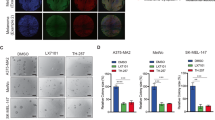
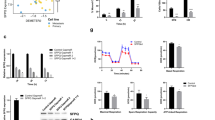
Melanoma is one of the most lethal forms of skin cancer because of its early metastatic spread. The variant form of CD44 (CD44v), a cell surface glycoprotein, is highly expressed on metastatic melanoma. The mechanisms of regulation of CD44 alternative splicing in melanoma and its pathogenic contributions are so far poorly understood. Here, we investigated the expression level of CD44 in a large set of melanocytic lesions at different stages. We found that the expression of CD44v8–10 and a splicing factor, U2AF2, is significantly increased during melanoma progression, whereas CD82/KAI1, a tetraspanin family of tumor suppressor, is reduced in metastatic melanoma. CD44v8–10 and U2AF2 expression levels, which are negatively correlated with CD82 levels, are markedly elevated in primary melanoma compared with dysplastic nevi and further increased in metastatic melanoma. We also showed that patients with higher CD44v8–10 and U2AF2 expression levels tended to have shorter survival. By using both in vivo and in vitro assays, we demonstrated that CD82 inhibits the production of CD44v8–10 on melanoma. Mechanistically, U2AF2 is a downstream target of CD82 and in malignant melanoma facilitates CD44v8–10 alternative splicing. U2AF2-mediated CD44 isoform switch is required for melanoma migration in vitro and lung and liver metastasis in vivo. Notably, overexpression of CD82 suppresses U2AF2 activity by inducing U2AF2 ubiquitination. In addition, our data suggested that enhancement of melanoma migration by U2AF2-dependent CD44v8–10 splicing is mediated by Src/focal adhesion kinase/RhoA activation and formation of stress fibers, as well as CD44-E-selectin binding reinforcement. These findings uncovered a hitherto unappreciated function of CD82 in severing the linkage between U2AF2-mediated CD44 alternative splicing and cancer aggressiveness, with potential prognostic and therapeutic implications in melanoma.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout










Similar content being viewed by others
References
Huh SJ, Liang S, Sharma A, Dong C, Robertson GP . Transiently entrapped circulating tumor cells interact with neutrophils to facilitate lung metastasis development. Cancer Res 2010; 70: 6071–6082.
Sharma A, Tran MA, Liang S, Sharma AK, Amin S, Smith CD et al. Targeting mitogen-activated protein kinase/extracellular signal-regulated kinase kinase in the mutant (V600E) B-Raf signaling cascade effectively inhibits melanoma lung metastases. Cancer Res 2006; 66: 8200–8209.
Zhang P, Goodrich C, Fu C, Dong C . Melanoma upregulates ICAM-1 expression on endothelial cells through engagement of tumor CD44 with endothelial E-selectin and activation of a PKCalpha-p38-SP-1 pathway. FASEB J 2014; 28: 4591–4609.
Zhang P, Ozdemir T, Chung CY, Robertson GP, Dong C . Sequential binding of alphaVbeta3 and ICAM-1 determines fibrin-mediated melanoma capture and stable adhesion to CD11b/CD18 on neutrophils. J Immunol 2011; 186: 242–254.
Zhang P, Fu C, Bai H, Song E, Song Y . CD44 variant, but not standard CD44 isoforms, mediate disassembly of endothelial VE-cadherin junction on metastatic melanoma cells. FEBS Lett 2014; 588: 4573–4582.
Zoller M . CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer 2011; 11: 254–267.
Prochazka L, Tesarik R, Turanek J . Regulation of alternative splicing of CD44 in cancer. Cell Signal 2014; 26: 2234–2239.
Ishii H, Saitoh M, Sakamoto K, Kondo T, Katoh R, Tanaka S et al. Epithelial splicing regulatory proteins 1 (ESRP1) and 2 (ESRP2) suppress cancer cell motility via different mechanisms. J Biol Chem 2014; 289: 27386–27399.
Reinke LM, Xu Y, Cheng C . Snail represses the splicing regulator epithelial splicing regulatory protein 1 to promote epithelial-mesenchymal transition. J Biol Chem 2012; 287: 36435–36442.
Brown RL, Reinke LM, Damerow MS, Perez D, Chodosh LA, Yang J et al. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J Clin Invest 2011; 121: 1064–1074.
Yae T, Tsuchihashi K, Ishimoto T, Motohara T, Yoshikawa M, Yoshida GJ et al. Alternative splicing of CD44 mRNA by ESRP1 enhances lung colonization of metastatic cancer cell. Nat Commun 2012; 3: 883.
Cheng C, Sharp PA . Regulation of CD44 alternative splicing by SRm160 and its potential role in tumor cell invasion. Mol Cell Biol 2006; 26: 362–370.
Takeo K, Kawai T, Nishida K, Masuda K, Teshima-Kondo S, Tanahashi T et al. Oxidative stress-induced alternative splicing of transformer 2beta (SFRS10) and CD44 pre-mRNAs in gastric epithelial cells. Am J Physiol Cell Physiol 2009; 297: C330–C338.
Tsai YC, Weissman AM . Dissecting the diverse functions of the metastasis suppressor CD82/KAI1. FEBS Lett 2011; 585: 3166–3173.
Charrin S, le Naour F, Silvie O, Milhiet PE, Boucheix C, Rubinstein E . Lateral organization of membrane proteins: tetraspanins spin their web. Biochem J 2009; 420: 133–154.
Berditchevski F, Odintsova E . Tetraspanins as regulators of protein trafficking. Traffic 2007; 8: 89–96.
Odintsova E, van Niel G, Conjeaud H, Raposo G, Iwamoto R, Mekada E et al. Metastasis suppressor tetraspanin CD82/KAI1 regulates ubiquitylation of epidermal growth factor receptor. J Biol Chem 2013; 288: 26323–26334.
Odintsova E, Sugiura T, Berditchevski F . Attenuation of EGF receptor signaling by a metastasis suppressor, the tetraspanin CD82/KAI-1. Curr Biol 2000; 10: 1009–1012.
Wei Q, Zhang F, Richardson MM, Roy NH, Rodgers W, Liu Y et al. CD82 restrains pathological angiogenesis by altering lipid raft clustering and CD44 trafficking in endothelial cells. Circulation 2014; 130: 1493–1504.
Jacobs PP, Sackstein R . CD44 and HCELL: preventing hematogenous metastasis at step 1. FEBS Letters 2011; 585: 3148–3158.
Nagano O, Okazaki S, Saya H . Redox regulation in stem-like cancer cells by CD44 variant isoforms. Oncogene 2013; 32: 5191–5198.
Konstantopoulos K, Thomas SN . Cancer cells in transit: the vascular interactions of tumor cells. Annu Rev Biomed Eng 2009; 11: 177–202.
Ponta H, Sherman L, Herrlich PA . CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol 2003; 4: 33–45.
Kim JH, Kim B, Cai L, Choi HJ, Ohgi KA, Tran C et al. Transcriptional regulation of a metastasis suppressor gene by Tip60 and beta-catenin complexes. Nature 2005; 434: 921–926.
Zhu S, Chen Z, Katsha A, Hong J, Belkhiri A, El-Rifai W . Regulation of CD44E by DARPP-32-dependent activation of SRp20 splicing factor in gastric tumorigenesis. Oncogene e-pub ahead of print 29 Jun 2015; doi:10.1038/onc.2015.250 2015.
Wei WJ, Mu SR, Heiner M, Fu X, Cao LJ, Gong XF et al. YB-1 binds to CAUC motifs and stimulates exon inclusion by enhancing the recruitment of U2AF to weak polypyrimidine tracts. Nucleic Acids Res 2012; 40: 8622–8636.
Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L et al. Widespread siRNA "off-target" transcript silencing mediated by seed region sequence complementarity. RNA 2006; 12: 1179–1187.
Stoilov P, Lin CH, Damoiseaux R, Nikolic J, Black DL . A high-throughput screening strategy identifies cardiotonic steroids as alternative splicing modulators. Proc Natl Acad Sci USA 2008; 105: 11218–11223.
Hanley WD, Burdick MM, Konstantopoulos K, Sackstein R . CD44 on LS174T colon carcinoma cells possesses E-selectin ligand activity. Cancer Res 2005; 65: 5812–5817.
Liang S, Sharma A, Peng HH, Robertson G, Dong C . Targeting mutant (V600E) B-Raf in melanoma interrupts immunoediting of leukocyte functions and melanoma extravasation. Cancer Res 2007; 67: 5814–5820.
McEver RP, Zhu C . Rolling cell adhesion. Annu Rev Cell Dev Biol 2010; 26: 363–396.
Wirtz D, Konstantopoulos K, Searson PC . The physics of cancer: the role of physical interactions and mechanical forces in metastasis. Nat Rev Cancer 2011; 11: 512–522.
Zeng Y, Wodzenski D, Gao D, Shiraishi T, Terada N, Li Y et al. Stress-response protein RBM3 attenuates the stem-like properties of prostate cancer cells by interfering with CD44 variant splicing. Cancer Res 2013; 73: 4123–4133.
Zhou B, Liu L, Reddivari M, Zhang XA . The palmitoylation of metastasis suppressor KAI1/CD82 is important for its motility- and invasiveness-inhibitory activity. Cancer Res 2004; 64: 7455–7463.
Sridhar SC, Miranti CK . Tetraspanin KAI1/CD82 suppresses invasion by inhibiting integrin-dependent crosstalk with c-Met receptor and Src kinases. Oncogene 2006; 25: 2367–2378.
Khanna P, Chung CY, Neves RI, Robertson GP, Dong C . CD82/KAI expression prevents IL-8-mediated endothelial gap formation in late-stage melanomas. Oncogene 2014; 33: 2898–2908.
Subbaram S, Lyons SP, Svenson KB, Hammond SL, McCabe LG, Chittur SV et al. Integrin alpha3beta1 controls mRNA splicing that determines Cox-2 mRNA stability in breast cancer cells. J Cell Sci 2014; 127: 1179–1189.
Mackereth CD, Madl T, Bonnal S, Simon B, Zanier K, Gasch A et al. Multi-domain conformational selection underlies pre-mRNA splicing regulation by U2AF. Nature 2011; 475: 408–411.
Marzese DM, Liu M, Huynh JL, Hirose H, Donovan NC, Huynh KT et al. Brain metastasis is predetermined in early stages of cutaneous melanoma by CD44v6 expression through epigenetic regulation of the spliceosome. Pigment Cell Melanoma Res 2015; 28: 82–93.
Wang X, Bruderer S, Rafi Z, Xue J, Milburn PJ, Kramer A et al. Phosphorylation of splicing factor SF1 on Ser20 by cGMP-dependent protein kinase regulates spliceosome assembly. EMBO J 1999; 18: 4549–4559.
Okamoto Y, Onogi H, Honda R, Yasuda H, Wakabayashi T, Nimura Y et al. cdc2 kinase-mediated phosphorylation of splicing factor SF2/ASF. Biochem Biophys Res Commun 1998; 249: 872–878.
Xiong F, Lin Y, Han Z, Shi G, Tian L, Wu X et al. Plk1-mediated phosphorylation of UAP56 regulates the stability of UAP56. Mol Biol Rep 2012; 39: 1935–1942.
Shirure VS, Liu T, Delgadillo LF, Cuckler CM, Tees DF, Benencia F et al. CD44 variant isoforms expressed by breast cancer cells are functional E-selectin ligands under flow conditions. Am J Physiol Cell Physiol 2015; 308: C68–C78.
Hanley WD, Napier SL, Burdick MM, Schnaar RL, Sackstein R, Konstantopoulos K . Variant isoforms of CD44 are P- and L-selectin ligands on colon carcinoma cells. FASEB J 2006; 20: 337–339.
Yago T, Shao BJ, Miner JJ, Yao LB, Klopocki AG, Maeda K et al. E-selectin engages PSGL-1 and CD44 through a common signaling pathway to induce integrin alpha(L)beta(2)-mediated slow leukocyte rolling. Blood 2010; 116: 485–494.
Takahashi K, Stamenkovic I, Cutler M, Saya H, Tanabe KK . CD44 hyaluronate binding influences growth kinetics and tumorigenicity of human colon carcinomas. Oncogene 1995; 11: 2223–2232.
Okamoto I, Morisaki T, Sasaki J, Miyake H, Matsumoto M, Suga M et al. Molecular detection of cancer cells by competitive reverse transcription-polymerase chain reaction analysis of specific CD44 variant RNAs. J Natl Cancer Inst 1998; 90: 307–315.
Konig H, Moll J, Ponta H, Herrlich P . Trans-acting factors regulate the expression of CD44 splice variants. EMBO J 1996; 15: 4030–4039.
van Weering DH, Baas PD, Bos JL . A PCR-based method for the analysis of human CD44 splice products. PCR Methods Appl 1993; 3: 100–106.
Zhang P, Feng S, Bai H, Zeng P, Chen F, Wu C et al. Polychlorinated biphenyl quinone induces endothelial barrier dysregulation by setting the cross talk between VE-cadherin, focal adhesion, and MAPK signaling. Am J Physiol Heart Circ Physiol 2015; 308: H1205–H1214.
Chesla SE, Selvaraj P, Zhu C . Measuring two-dimensional receptor-ligand binding kinetics by micropipette. Biophys J 1998; 75: 1553–1572.
Fu C, Tong C, Wang M, Gao Y, Zhang Y, Lu S et al. Determining beta2-integrin and intercellular adhesion molecule 1 binding kinetics in tumor cell adhesion to leukocytes and endothelial cells by a gas-driven micropipette assay. J Biol Chem 2011; 286: 34777–34787.
Acknowledgements
We thank Go J Yoshida (Tokyo Medical and Dental University) for helpful discussion. This study was funded by Fundamental Research Funds for the Central Universities (XDJK2014C176 to PZ, XDJK2012B010 to HFZ, XDJK2015C154 to QR and XDJK2015C155 to FS), the Start-up Foundation of Southwest University (SWU114017 to PZ), National Natural Science Foundation of China (NSFC) (NSFC-81402393 to PZ, NSFC-81572678 to PZ, NSFC-81272433 to GTL, NSFC-81472732 to GTL and NSFC-81073084 to HFZ) and National Science Foundation grants (CBET-0729091 to CD).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Zhang, P., Feng, S., Liu, G. et al. CD82 suppresses CD44 alternative splicing-dependent melanoma metastasis by mediating U2AF2 ubiquitination and degradation. Oncogene 35, 5056–5069 (2016). https://doi.org/10.1038/onc.2016.67
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.67
This article is cited by
-
TGF-β1 promotes epithelial-to-mesenchymal transition and stemness of prostate cancer cells by inducing PCBP1 degradation and alternative splicing of CD44
Cellular and Molecular Life Sciences (2021)
-
The U2AF2 /circRNA ARF1/miR-342–3p/ISL2 feedback loop regulates angiogenesis in glioma stem cells
Journal of Experimental & Clinical Cancer Research (2020)
-
hnRNPK promotes gastric tumorigenesis through regulating CD44E alternative splicing
Cancer Cell International (2019)
-
The impact of the RBM4-initiated splicing cascade on modulating the carcinogenic signature of colorectal cancer cells
Scientific Reports (2017)