Abstract
B-cell receptor (BCR) signaling is essential for the development of B cells and has a critical role in B-cell neoplasia. Increasing evidence indicates an association between chronic hepatitis C virus (HCV) infection and B-cell lymphoma, however, the mechanisms by which HCV causes B-cell lymphoproliferative disorder are still unclear. Herein, we demonstrate the expression of HCV viral proteins in B cells of HCV-infected patients and show that HCV upregulates BCR signaling in human primary B cells. HCV nonstructural protein NS3/4A interacts with CHK2 and downregulates its activity, modulating HuR posttranscriptional regulation of a network of target mRNAs associated with B-cell lymphoproliferative disorders. Interestingly, the BCR signaling pathway was found to have the largest number of transcripts with increased association with HuR and was upregulated by NS3/4A. Our study reveals a previously unidentified role of NS3/4A in regulation of host BCR signaling during HCV infection, contributing to a better understanding of the molecular mechanisms underlying HCV-associated B-cell lymphoproliferative disorders.
Similar content being viewed by others
Introduction
Hepatitis C virus (HCV) is an enveloped, positive-strand RNA virus belonging to the Flaviviridae family. Complications of chronic HCV infection include cirrhosis, decompensated liver disease and hepatocellular carcinoma. Extrahepatic diseases such as mixed cryoglobulinemia and B-cell non-Hodgkin's lymphoma (B-cell NHL) are often identified in patients with chronic HCV. There are three lines of evidence supporting an association between HCV and B-NHL. First, epidemiological data indicate a strong link between persistent HCV infection and B-cell NHL.1 Second, clinical data have shown that antiviral therapy resulted in remissions of lymphoma in HCV-positive but not HCV-negative NHL patients.2 Third, experimental data demonstrate that transgenic mice expressing the full-length HCV genome specifically in B cells had a higher incidence of B-cell NHL, primarily diffuse large B-cell lymphoma (DLBCL).3 HCV does not contain an obvious oncogene and does not integrate into host genomes. The mechanisms by which HCV infection causes B-cell lymphoma remain elusive. Understanding the mechanism may contribute to identification of newer drug targets for HCV-associated lymphoproliferative disorders.
The HCV RNA genome encodes a single long open reading frame, which is processed by host and viral proteases into at least three structural and seven nonstructural proteins in the following order: core, envelope 1 (E1), E2, p7, nonstructural 2 (NS2), NS3, NS4A, NS4B, NS5A and NS5B. NS4A binds NS3 and functions as a cofactor for both the serine protease and RNA helicase activities of the NS3 enzyme.4 NS3/4A is known to modulate the host antiviral immune system by protein cleavage.5, 6 It has been reported that HCV NS3/4A protein interacts with ATM (ataxia telangiectasia mutated) and impairs DNA repair in non-lymphoid cells.7 Checkpoint kinase 2 (CHK2) is one of the key downstream molecules of ATM. Given the possible link between HCV NS3/4A and CHK2, we hypothesize that CHK2 signaling may be modulated by HCV infection.
B-cell receptor (BCR) signaling is critical for the development of normal B cells and B-cell lymphoma.8 The BCR includes membrane immunoglobulin molecules and associated CD79A/CD79B (Igα/Igβ) heterodimers. Antigen binds to the surface immunoglobulin of the BCR and induces BCR aggregation. Antigen-induced BCR aggregation elicits Src-family kinases to phosphorylate CD79A/CD79B and subsequently phosphorylates the tyrosine kinase SYK. SYK activation triggers a signaling cascade that includes the tyrosine kinases Bruton’s tyrosine kinase (BTK) and CARD11.9 It is unclear whether the BCR signaling pathway is involved in HCV-associated B-cell lymphoproliferative disorders.
Many studies have demonstrated HCV infection of peripheral blood B cells of chronic HCV patients using polymerase chain reaction (PCR)-based methods,10, 11, 12, 13 although some studies have shown conflicting results.14, 15, 16 HCV core and NS3 have been detected in CD19+ but not CD19– peripheral blood mononuclear cells by real-time reverse transcriptase (RT)–PCR, immunoblot analysis and enzyme immunoassay.12 HCV has been shown to infect B cells both in vitro and in vivo.10, 17, 18, 19, 20
In this study, we demonstrate HCV infection in the B-cell subset of chronic hepatitis C patients and B cells exposed ex vivo to authentic patient-derived HCV, and find that these HCV-infected B cells have upregulated BCR signaling. These results underscore a putative relationship between HCV infection and B-cell lymphomagenesis. Furthermore, our results establish a hierarchy of molecular events in which NS3/4A overexpression interferes with CHK2 activity, which in turn leads to alteration of HuR activity and subsequent posttranscriptional modulation of its target mRNAs. The BCR signaling pathway was the top-ranked pathway showing increased association with HuR and upregulated by NS3/4A overexpression. Our findings highlight a critical biological role of NS3/4A in the regulation of BCR signaling during HCV infection and contribute to a better understanding of the molecular mechanisms underlying HCV infection.
Results
Detection of HCV viral proteins in peripheral blood B cells in HCV-infected patients
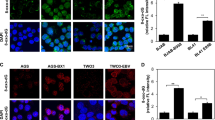
A number of groups have detected HCV RNA in B cells of HCV-infected patients using PCR-based methods.10, 11, 12, 13 To examine the expression of HCV viral proteins in B cells, we purified B cells from human normal and HCV-infected peripheral blood mononuclear cells (five and six patients, respectively) (Supplementary Table S1), and conducted double-immunofluorescence analysis of B-cell markers (CD19, CD20 and B220) and viral proteins (NS3 and core) in these cells. Although B220 is a pan B-cell marker in mice, it is expressed in a subset of human B cells that do not express CD27.21 The specificity of these antibodies was validated and described in Supplementary Figure S1. HCV viral proteins were detected in the B cells of human HCV-infected patients (Figures 1a–c). HCV core protein was also observed in some B220-positive cells (Figure 1d). The percentage of NS3-positive cells and core-positive cells detected in HCV-infected CD19+ B cells by double-immunofluorescence analysis was 68% and 38%, respectively. The percentage of both CD19 and NS3-positive cells was determined by flow cytometry with fluorescein isothiocyanate anti-human CD19 antibody and Alexa Fluor 647 anti-HCV NS3 antibody, as well as appropriate isotype controls. As shown in Supplementary Figure S2, 80.1% of CD19-positive cells are NS3 positive. This percentage is a little bit higher than that detected by double-immunofluorescence staining, which may be due to the higher sensitivity of flow cytometry. The susceptibility of B cells to HCV was verified ex vivo by infection of primary B cells purified from healthy donors and Raji cells using authentic virus from chronically infected patients (patients 10–12; Supplementary Table S1) and detection of HCV RNA-positive and -negative (replicative) strands (Supplementary Figures S3A and B) and viral core protein (Supplementary Figure S4). Together, these results further support HCV infection in B cells.
Detection of HCV viral proteins in B cells of HCV-infected patients. Representative images at × 600 magnification were captured by confocal immunofluorescence microscopy in human normal B cells and HCV-infected B cells. Magnified insets with representative single cells were shown on the left corner of each image for further clarification. DAPI, 4′,6-diamidino-2-phenylindole. (a) NS3 (red) and CD19 (green) staining. Merge: merged image of CD19, NS3 and DAPI. (b) HCV core (red) and CD19 (green) staining. Merge: merged image of CD19, core and DAPI. (c) HCV core (red) and CD20 (green) staining. Merge: merged image of CD20, core and DAPI. (d) HCV core (red) and B220 (green) staining. Merge: merged image of B220, core and DAPI.
HCV NS3/4A downregulates CHK2 activity in B-cell lymphoma
The molecular mechanism by which HCV infection leads to B-cell lymphoma remains speculative. It has been reported that HCV NS3/4A protein interacts with ATM and impairs DNA repair in non-lymphoid cells.7 The CHK2 kinase is a key sensor to DNA damage. In order to study whether NS3/4A interferes with the CHK2 pathway in HCV-infected lymphoma, we first examined whether CHK2 can be cleaved by NS3/4A, as NS3/4A is a known protease. Our results show that there are no cleavage products of CHK2 in the presence of NS3/4A, indicating that CHK2 is not a substrate of NS3/4A protease (Figure 2a). We next conducted double-immunofluorescence analysis of NS3 and CHK2 in human HCV-infected or normal B cells to evaluate their colocalization. As shown in Figure 2b, NS3 and CHK2 were predominately colocalized in the cytoplasm of human HCV-infected B cells. Physical association of NS3/4A to CHK2 was therefore analyzed by immunoprecipitation (IP). Interestingly, we were able to detect IP of endogenous CHK2 with NS3/4A in SUDHL6 cells stably transduced with NS3/4A expression plasmids (Figure 2c). To further confirm this interaction and to demonstrate a direct physical association, in vitro glutathione S-transferase (GST) fusion protein pull-down assays were performed. We showed that the proteins from GST-CHK2 wild type, truncated forms and T68A mutant were able to pull-down NS3/4A (Figure 2d), demonstrating that there is indeed a physical interaction between CHK2 and NS3/4A. To further determine which domain of NS3/4A protein is critical for CHK2–NS3/4A interaction, we expressed a series of FLAG epitope-tagged NS3/4A truncation and deletion mutants in HEK and HeLa cells, due to the technical difficulty in transfecting DNA into SUDHL6 cells using liposomal or electroporation methods. Co-IP analysis revealed that the protease domain of NS3/4A protein is critical for CHK2 binding with NS3/4A but the protease activity is not required for their interaction in HeLa and HEK cells (Supplementary Figure S5). To further confirm this in SUDHL6 cells, lentiviruses encoding full-length NS3/4A, protease domain, helicase domain and its proteolytically inactive form S139A mutant protein were generated. As shown in Figure 2e, the protease domain of NS3/4A protein, but not the protease activity, is required for CHK2–NS3/4A interaction in SUDHL6 cells, which is consistent with results in HeLa and HEK cells.
HCV NS3/4A interacts with CHK2 and downregulates its activity in lymphoma cells. (a) CHK2 is not a substrate for NS3/4A protease. SUDHL6 and SUDHL4 cells were stably transduced with lentivirus encoding N3/4A or vector control (V) and the lysates were subjected to western blot analysis for CHK2 and NS3. Actin was used as a loading control. (b) Colocalization of CHK2 and NS3 in human normal B cells and HCV-infected B cells. Double immunofluorescent images for CHK2 (green) and NS3 (red) by confocal immunofluorescence microscopy at × 600 magnification indicates that there is a strong overlap in human HCV-infected B cells. DAPI, 4′,6-diamidino-2-phenylindole; Merge, merged image of CHK2, NS3 and DAPI. (c) NS3/4A associates with CHK2 in NS3/4A stably transduced SUDHL6 cells. SUDHL6-NS3/4A cells were IP using anti-CHK2 antibody, followed by immunoblotting (IB) with NS3 and CHK2. Expression of CHK2 and NS3 was monitored by immunoblotting of total cell lysates. Vector control (V) and SUDHL6-NS3/4A cells (two clones; 2 and 3) were analyzed, actin was used as a loading control. (d) NS3/4A associates with CHK2 in vitro. GST-fused CHK2 truncations and wild-type (WT) proteins were immobilized onto glutathione sepharose 4B beads and incubated with SUDHL6-NS3/4A cell lysates (three clones; 1, 2 and 3). Immunoblotting using anti-NS3 antibody was performed. GST fusion proteins were visualized with Coomassie blue staining (CBS). (e) The protease domain of NS3/4A is critical for the interaction between NS3/4A and CHK2 proteins. The SUDHL6 cells were transduced with lentiviruses encoding FLAG-tagged full-length NS3/4A, proteinase domain, helicase domain and S139A mutant protein. The transduced cells were IP using anti-FLAG antibody, followed by immunoblotting with CHK2 and FLAG. (f) Overexpression of NS3/4A downregulates CHK2 activity in SUDHL6 cells. SUDHL6-NS3/4A (Mass: mass culture, or 1, 2, 3: clones) or vector control cells (V) were lysed and the lysates were subjected to western blot analysis using CHK2, anti-pCHK2 T68, p53, and anti-p53 Ser20 antibodies. (g) Densitometric analysis showing relative band intensity of phospho-CHK2 Thr68 in SUDHL6-NS3/4A (Mass: mass culture, or 1, 2, 3: clones) or vector control cells as represented by western blot in f (mean±s.d., n=3, *P-value <0.05), normalized to actin (set as value 1).
The interaction of CHK2 and NS3/4A suggests that they may regulate each other. This was supported by our observation that NS3/4A downregulated CHK2 phosphorylation at Thr68 (Figures 2f and g). Phosphorylation of CHK2 at Thr68 functions as a surrogate marker for CHK2 activation,22 therefore our data indicate that NS3/4A downregulates CHK2 activity. Phosphorylation of p53, a downstream molecule of CHK2, at serine 20, is also inhibited by NS3/4A overexpression (Figures 2f and g), further indicating downregulation of CHK2 activity. NS3/4A physically interacts with CHK2 while downregulating CHK2 activity, consistent with inhibition of DNA damage repair by NS3/4A.
Differential association of HuR with target mRNAs in NS3/4A-overexpressing SUDHL6 cells versus control cells
CHK2 is known to interact with and phosphorylate HuR, whereas HuR subsequently regulates many mRNAs that encode proteins involved in stress response, cell proliferation and tumorigenesis.23, 24 We aimed to examine the influence of CHK2 activity on global HuR posttranscriptional gene regulation in response to NS3/4A expression. To gain insight into the alteration of HuR-binding activity caused by the overexpression of NS3/4A, we performed ribonucleoprotein IP analysis using an anti-HuR antibody followed by microarray analysis of the obtained mRNAs, and then compared the collection of mRNAs associated with HuR in control cells and in NS3/4A-overexpressing SUDHL6 cells. The association of mRNAs with HuR was assessed in three independent experiments (Supplementary Figure S6) by measuring the enrichment of mRNAs in HuR IP samples of SUDHL6-NS3/4A cells compared with control cells. Venn diagram analysis of the common and specific genes associated with HuR showed that 200 of 1324 genes did not have an altered association with HuR, whereas 534 genes were upregulated and 590 genes were downregulated in NS3/4A-overexpressing cells compared with control cells (Figure 3a). Among the top 50 genes exhibiting significant changes in association with HuR under NS3/4A overexpression were two major components of BCR signaling, CD79A and CD79B (Figure 3b), which are critical for B-cell development and growth. These data demonstrated that NS3/4A was involved in regulating gene expression posttranscriptionally by modulating the association of HuR with its target mRNAs. As NS3 mRNA is not included in the gene lists of standard human microarray chips, we next examined whether NS3 mRNA binds to HuR. Interestingly, we found that NS3 mRNA was associated with HuR (Supplementary Figure S7).
Differential association of HuR with target mRNAs in NS3/4A-overexpressing SUDHL6 cells versus control cells. (a) Venn diagram analysis shows the number of genes upregulated (red) or downregulated (green) associated with HuR in SUDHL6-NS3/4A cells relative to control cells. (Z-ratio differences ⩾1.5 or⩽−1.5, P-value <0.05). (b) Heatmap image demonstrates the top 50 genes with the largest changes in association with HuR in NS3/4A-overexpressing cells relative to control. The difference in association with HuR was measured by Z-ratio values, increasing from blue to red.
Functional analysis of the mRNAs differentially associated with HuR upon NS3/4A overexpression
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway databases were used to analyze the microarray data in order to examine whether the genes differentially associated with HuR in NS3/4A overexpression are of functional importance. Analysis revealed that the BCR signaling pathway was characterized by the largest number of transcripts with increased association with HuR in response to NS3/4A overexpression (Figure 4a). The top five pathways affected by NS3/4A overexpression included those related to B-cell development and proliferation. To understand the potential functional implications of the transcripts on HuR-mRNA association, genes markedly affected by NS3/4A overexpression were placed in the context of the known molecular interactions using ingenuity pathway analysis. Genes related to the top-ranked network, the BCR signaling pathway, demonstrated altered association with HuR in response to NS3/4A overexpression. The BCR signaling pathway was centered on the increase in HuR binding to CD79A mRNA (Figure 4b), surrounded by other interacting genes whose association with HuR was affected by NS3/4A overexpression. The ingenuity pathway analysis data further supports the notion that NS3/4A overexpression preferentially upregulates the BCR signaling pathway centered on CD79A.
The BCR signaling pathway is the top-ranked network with increasing association with HuR in response to NS3/4A overexpression. (a) KEGG pathway analysis revealed that the BCR signaling pathway has the largest number of transcripts with increasing association with HuR in response to NS3/4A overexpression. Red and pink represent upregulated genes; and green, downregulated genes. (b) The top-ranked network of genes was centered on the increase in HuR binding to CD79A mRNA, surrounded by other interacting genes whose association with HuR was affected by NS3/4A overexpression.
Analysis of HuR targets and upregulation of BCR signaling by NS3/4A
To further corroborate the changes of the transcripts showing differential association with HuR in NS3/4A overexpression by microarray, we selected four targets of the BCR signaling pathway for detailed verification using real-time quantitative reverse transcription PCR (RT–qPCR). All of the selected genes have critical roles in regulation of B-cell development and B-cell lymphomagenesis. CARD11, Blk, SYK and BCL6 mRNA in association with HuR were upregulated in NS3/4A-overexpressing cells as compared with control cells, confirming microarray data (Figure 5a). In addition, we also selected two downregulated genes (ALDH1A1 and ALDH1A2) for comparison, as their associations with HuR were decreased in NS3/4A-overexpressing cells. ALDH1A1 expression was markedly reduced in non-small cell lung carcinomas.25 Touma et al.26 demonstrated lower cytoplasmic ALDH1A2 protein levels in all human prostate cancer sections relative to normal human prostate tissue. Consistent with the microarray result, both ALDH1A1 and ALDH1A2 mRNA in association with HuR were decreased in NS3/4A-overexpressing cells (Figure 5a). As the targets of the BCR signaling pathway in association with HuR were upregulated, whether HuR regulated its target mRNA stability was subsequently investigated. Actinomycin D was used to treat vector control and NS3/4A-overexpressing SUDHL6 cells in order to inhibit new transcription. Overexpression of NS3/4A increased the half-life of Blk mRNA from 5.5 to 9.5 h and the half-life of SYK mRNA from 5.5 to 9 h in SUDHL6 cells (Supplementary Figure S8), indicating that the mRNA-binding protein HuR may have an important role in Blk and SYK mRNA stability.
Analysis of HuR targets modulated by NS3/4A overexpression. (a) The levels of selected HuR target mRNAs in control and SUDHL6-NS3/4A (mass culture) cells were detected by ribonucleoprotein (RNP)-IP with anti-HuR antibody, followed by RT–qPCR analysis. Data show mean and s.d. from at least three separate experiments. * and ** indicate values significantly greater (*P<0.05, and **P<0.01) in SUDHL6-NS3/4A cells (black column) over control cells (white column). (b) Western blot analysis showing protein expression of selected HuR targets in whole cell lysates of vector control (V) and SUDHL6-NS3/4A (NS3/4A) cells. Actin was used as a loading control. (c) Telaprevir treatment attenuates the upregulation of CD79A and CD79B protein levels by NS3/4A. SUDHL6 vector control and NS3/4A-overexpressing cells were treated with 10 μM Telaprevir and its vehicle control (DMSO) for 24 h. Protein level of CD79A, CD79B and NS3 was examined by western blot analysis.
Next, the proteins encoded by these mRNAs were assessed by western blot analysis. BCR components CD79A and CD79B were more abundant in NS3/4A-overexpressing cells and protein expression of proximal BCR signaling effectors SYK, BTK, Blk, BCL6 and CARD11 were markedly increased in NS3/4A-overexpressing SUDHL6 cells compared with control cells (Figure 5b). Similar results were observed in control and NS3/4A-overexpressing SUDHL2 cells (Supplementary Figure S9). These data together indicate that the BCR signaling pathway was preferentially upregulated by the NS3/4A-CHK2-HuR cascade. As NS3/4A is a protease, we wonder whether the protease function of NS3/4A is involved in the upregulation of HuR-regulated transcripts. Protease inhibitor (Telaprevir) treatment attenuated upregulation of CD79A and CD79B protein levels by NS3/4A (Figure 5c), indicating that protease function of NS3/4A may be required for its upregulation of HuR-regulated transcripts.
As NS3/4A upregulated the protein levels of the BCR signaling pathway, we asked whether NS3/4A could change global protein synthesis. We analyzed the ribosome-RNA engagement in polysome and non-polysome fractions from both SUDHL6-NS3/4A cells and control cells. As shown in Supplementary Figure S10, polysome fractions of NS3/4A-expressing cells consistently exhibited a similar profile as that of control cells, indicating that NS3/4A did not alter the pattern of global protein synthesis, but preferentially regulated the translation of a specific set of transcripts, especially those associated with BCR signaling.
HCV infection upregulates BCR signaling in primary human B cells and Raji cells
As NS3/4A upregulated BCR signaling, we next asked whether HCV infection induces the upregulation of BCR signaling in primary human B cells. Owing to limited numbers of purified B cells from individual HCV patients, we pooled cells from eight different HCV-infected patients in order to isolate a sufficient quantity of protein for analysis. As shown in Figure 6a, the protein levels of CD79A and CARD11 were markedly increased in HCV-infected B cells compared with normal B cells in primary blood samples. In HCV-infected Raji cells, the protein expression of both HCV NS3 and core was detected by western blot (Figure 6b). In addition, two major components (CD79A and CD79B) of the BCR are upregulated in HCV-infected Raji cells compared with control (Figure 6b). These experiments further validated that BCR signaling was upregulated by HCV infection ex vivo, in line with HCV NS3 expression. These data indicated that HCV upregulates the host BCR signaling pathway at least partly through its protein NS3/4A, which may lead to B-cell lymphoma of HCV-infected patients.
HCV infection upregulates BCR signaling in human primary B cells and Raji cells. (a) Western blot analysis showing protein expression of NS3, CARD11, Blk and CD79A in lysates isolated from normal (five discrete patients) and HCV-infected (mix: pooled from eight patients) purified B cells. Actin was used as a loading control. (b) Protein expression of NS3, core, CD79A and CD79B in lysates isolated from control and HCV-infected Raji cells was detected by western blot analysis. (c) The model depicts a proposed mechanism for HCV-associated B-cell lymphoproliferative disorders. HCV NS3/4A interacts with CHK2 and downregulates CHK2 activity. Subsequently, this repressed CHK2 activity modulates HuR posttranscriptional regulation of a network of target mRNAs associated with B-cell lymphoproliferative disorders, preferentially those involved in the BCR signaling pathway.
Discussion
Although a growing number of studies have detected the expression of HCV viral proteins in infected B cells,10, 12, 27 whether HCV infects B cells is still not universally accepted. It has been suggested that viral material in B cells is taken up from HCV present in plasma. To exclude this possibility, Ito et al.12 have provided compelling evidence that HCV preferentially infects peripheral B cells in chronic HCV patients by using semiquantitative real-time PCR, immunoblot analysis and enzyme immunoassays. Corroborative evidence to support HCV infection of primary B cells in vivo was provided by the Lai lab (Taipei, Taiwan), who successfully established EBV-immortalized B-lymphoid cell lines from HCV-infected patients and detected HCV RNA persistently during 3 months of continuous cell culture.17 In this study, we successfully demonstrated that authentic, patient-derived HCV infects normal human primary B cells and Raji cells ex vivo. We also demonstrated the expression of HCV proteins in primary human HCV-infected B cells by confocal microscopy, further confirming that HCV infects B cells. Longitudinal experiments are necessary to establish whether these core and CD19 double-positive cells actually give rise to B-cell lymphoma in vivo. Ng et al.28 have recently shown that the rescued lymphoma BCRs have no reactivity with HCV viral proteins and particles, therefore they conclude that most lymphomas of HCV-infected patients do not arise from B cells aimed at eliminating the virus. Their study was performed in lymphoma cells, rather than in HCV-infected non-malignant lymphocytes. It is plausible that HCV potentially transforms cells by a ‘hit and run’ mechanism. We propose a model in which HCV may initially stimulate B-cell proliferation; subsequently, the transformed B cells no longer require continuous HCV stimulation and their BCRs may have undetectable reactivity with HCV. Our study has not directly addressed this issue. However, we have provided evidence that HCV can activate the BCR signaling pathway by infection of B cells as depicted in Figure 6c. Machida et al.18 demonstrated that HCV infection in B cells induced mutations of cellular genes, which may contribute to B-cell lymphomagenesis.
The molecular mechanism by which HCV infection leads to B-cell lymphoma has not yet been definitively resolved. Our results show that NS3/4A downregulates CHK2 activity and its downstream p53 phosphorylation at serine 20. CHK2 kinase has a key role in cellular DNA damage response.29 CHK2 stimulates expression of DNA repair genes by stabilizing the FoxM1 transcription factor.30 It has been previously reported that HCV core protein inhibits the formation of the Mre11/NBS1/Rad50 complex, causing inhibition of DNA repair in monocytes and hepatocytes.31 Similarly, another study has also shown that NS3 protein directly binds to ATM and inhibits DNA repair in non-lymphoid cells.7 The inhibition of DNA damage repair makes HCV-infected cells more prone to mutations and potentiates chromosome instability.31 Our data reveal NS3/4A-mediated downregulation of CHK2 activity as an additional novel mechanism for DNA damage repair inhibition by HCV.
We have previously shown that HuR functions as a downstream effector of ATM/CHK2 phosphorylation, enhancing its association with specific subsets of mRNAs while reducing its association with others.32 Herein, we used IPs of ribonucleoprotein complexes followed by microarray analysis (RIP-Chip) to examine the differential associations of HuR with subsets of target mRNAs driven by overexpression of NS3/4A. Significantly, we identified a network of functionally interacting proteins encoded by mRNAs differentially bound to HuR in response to NS3/4A overexpression. Phosphorylation at each of the three HuR substrate residues (Ser88, Ser100 and Thr118) had a distinct influence on the association of HuR with its targets.32, 33 Our previous study has shown that in response to ionizing radiation exposure, HuR associates with target mRNAs when Ser100 is phosphorylated and dissociates when Thr118 is phosphorylated.32 All extant data suggest that the HuR phosphorylation site is very selective under different stimuli. Future work will be needed to identify which HuR residue is phosphorylated by CHK2 in response to NS3 overexpression.
Although it is well known that HuR regulates numerous genes posttranscriptionally, it is of great interest to identify specific genes/pathways that are selectively altered in HCV infection of B cells through the NS3/4A-CHK2-HuR regulation cascade. We have demonstrated that the BCR signaling pathway is among the most affected pathways with the largest number of transcripts showing increased association with HuR in response to NS3/4A overexpression. BCR signaling is essential for the survival of normal B cells. ‘Chronic active’ BCR signaling is required for cell survival in the activated B-cell-like (ABC) subtype of DLBCL.9 In our study, protein expression of BCR components CD79A and CD79B was significantly upregulated in response to NS3/4A overexpression in both SUDHL2 and SUDHL6 cells. CD79A-CD79B heterodimer is a key scaffold for mediating BCR signaling and initiation of downstream signaling to the nuclear factor-κB, phosphatidylinositol 3 kinase and mitogen-activated protein kinase pathways, which promote proliferation and survival of normal and malignant B cells. CARD11 is required for the activation of nuclear factor-κB signaling and survival of ABC DLBCL cells.9 Knockdown of BTK by its specific short hairpin RNA killed ABC DLBCL cells with wild-type CARD11.9 We have demonstrated that CARD11 and BTK are upregulated in response to either NS3/4A overexpression or HCV chronic infection. HCV may preferentially manipulate the host BCR signaling pathway at least partly through its protein NS3/4A, which may lead to B-cell lymphoproliferative disorders.
In summary, we demonstrated the expression of HCV viral proteins in primary B cells from patients infected with HCV. Furthermore, we established that HCV NS3/4A downregulates CHK2 activity to inhibit DNA damage repair. The BCR signaling pathway was characterized by the largest number of transcripts with increased association with HuR and upregulated by NS3/4A. The findings presented here highlight a novel and critical biological role of NS3/4A in regulation of BCR signaling during HCV infection and reveal a putative molecular mechanism underlying HCV-associated B-cell lymphoma. NS3/4A-induced upregulation of host BCR signaling may contribute to B-cell lymphomagenesis in HCV-infected patients.
Materials and methods
Cell culture and transduction
DLBCL (SUDHL2, SUDHL4 and SUDHL6) and Raji (CCL-86; ATCC, Manassas, VA, USA) cells were cultured in RPMI-1640 (Invitrogen, Grand Island, NY, USA) containing 10% fetal bovine serum. Cell lines were obtained from ATCC and confirmed free of mycoplasma contamination. For transduction of SUDHL6 cells, plasmids pCDH-Vector or pCDH-NS3/4A (System Biosciences, Mountain View, CA, USA) were transfected with PureFection (System Biosciences) in HEK-293TN packaging cells to generate lentivirus. SUDHL6 cells were co-cultured with the lentivirus encoding NS3/4A or control for 72 h and selected with 0.5 mg/ml puromycin (Invitrogen) for 14 days before being expanded for further experimentation.
A series of FLAG epitope-tagged NS3/4A truncation and deletion mutants were a generous gift from Dr Michael Gale Jr (University of Washington, WA, USA) and were transfected with PureFection in HEK and HeLa cells. At 48-h post-transfection, cells were harvested for analysis. The stable cell lines of SUDHL6 overexpressing FLAG epitope-tagged NS3/4A truncation and deletion mutants were generated by lentiviral transduction and were selected and maintained with 0.5 mg/ml puromycin. The expression lentiviral constructs overexpressing FLAG epitope-tagged NS3/4A truncation and deletion mutants were made as follows. FLAG-tagged NS3/4A truncation and deletion mutants were cloned by PCR (with primers containing a 5' EcoRI site and 3' BamHI site) into a lentiviral-vector CD532 (System Biosciences).
For the NS3/4A overexpression in SUDHL2 cells, the SUDHL2 cell line (CRL-2956, ATCC) was transduced with a lentivirus containing the pLenti-CMV-rtTA3-Blast plasmid (Addgene #26429, Cambridge, MA, USA) to generate cells, which constitutively express the tetracycline responsive transactivator. Cells were treated with blasticidine (Invitrogen) for a minimum of 3 weeks to generate a stable cell line. The NS3/4A gene fusion was amplified by PCR using the following primers: 5'-AGCTGGATCCGCCACCATGGCGCCTATTACGGCCTACTC-3' and 5'-AGCTTCTAGAAGCTACGCGTTTAGCACTCTTCCATCTCATCGAACTCC-3'. The PCR product was cloned into pLenti-CMVTRE3G (Addgene #27568) via BamHI/XbaI (ThermoFisher Scientific, Waltham, MA, USA) digest to generate pLenti-CMVTRE3G-NS3/4A. Empty vector was created by digesting with BamHI/XbaI, blunting with T4 polymerase (ThermoFisher Scientific) and ligating. Following sequence verification, lentivirus was produced and purified using the NS3/4A and empty vector constructs. Both viruses were then used to transduce the rtTA3G-stable SUDHL2 cell line, and selected with puromycin.
B-cell preparation and infection with HCV
Human primary sample studies have been approved by the University of Maryland Medical School Institutional Review Board or the Health Research Ethics Authority at Memorial University, Canada, and conform to the Declaration of Helsinki. Consent was obtained for the primary samples as required by the above noted authorities. Normal B cells were purified from healthy human donor peripheral blood mononuclear cells purchased from Astarte Biologics, LLC (Bothell, WA, USA) or acquired from volunteers. HCV-infected B cells were purified from peripheral blood mononuclear cells obtained from 14 patients with chronic HCV infection, as demonstrated by high plasma loads of HCV RNA (Supplementary Table S1). All patients were negative for HIV infection. B-cell isolation was performed using human B-Cell Isolation Kit (B-CLL; Miltenyi Biotec Inc., San Diego, CA, USA) according to the manufacturer’s protocol. Purity of CD19+/CD3− cells was determined as >95% by flow cytometry. Primary B cells from two healthy donors and Raji cells were infected with authentic HCV contained in plasma of treatment-naive individuals chronically infected with HCV (patients 10–12; Supplementary Table S1). For primary cells, 5 × 106 cells in 5 ml of RPMI-1640 with 10% fetal bovine serum, 2 mM/ml glutamine, 50 U/ml penicillin, 50 μg/ml streptomycin, 0.1 mM nonessential amino acids (Invitrogen) and 1 ng/ml of human interleukin-4 (IL-4; Roche Molecular Diagnostics, Pleasanton, CA, USA) were supplemented with HCV from patient 10 at 1:1 virus:cell ratio, incubated for 24 h, washed twice and cultured for 3 days. For Raji cells, 3.5 × 105 cells in 3 ml of the above medium without IL-4 were supplemented with HCV from patients 10, 11 or 12 at 1:1 virus:cell ratio and cultured for 3 days. After culture, both primary and Raji cells were extensively washed, counted and either RNA extracted or cells were fixed in 4% paraformaldehyde for immunostaining and image analyses. In all infection experiments, primary B cells and Raji cells not exposed to HCV (uninfected) and incubated with normal human plasma under the same culture conditions as the cells exposed to HCV were included as negative controls.
Detection of HCV RNA-positive and -negative strands
HCV RNA-positive (vegetative) strand was detected by RT–PCR with signal detection by nucleic acid hybridization using primers specific for the HCV 5’-untranslated region; cycling conditions, standards and controls reported previously.34 For identification of HCV RNA-negative (replicative) strand, RT–PCR/nucleic acid hybridization with rTth DNA polymerase performed under the negative strand-specific amplification conditions was applied, as detailed before.34, 35 In every analysis, a number of negative, contamination and positive controls were included, as shown in Supplementary Figure S3.
Detection of NS3-positive cells by flow cytometry
Mouse monoclonal anti-HCV NS3 antibody (Abcam, Cambridge, MA, USA; ab65407) was conjugated with Alexa Fluor 647 dye using Alex Fluor 647 Monoclonal Antibody Labeling Kit (Invitrogen; #A20186)) according to the manufacturer’s protocol. To determine the percentage of NS3-positive cells among CD19-positive cells, B cells were isolated using human B-Cell Isolation Kit and stained with fluorescein isothiocyanate anti-human CD19 antibody (Biolegend, San Diego, CA, USA; #302205) and Alexa Fluor 647 anti-HCV NS3 antibody, as well as appropriate isotype controls. The percentage of positive cells was analyzed by flow cytometry.
Immunofluorescence using confocal microscopy
Double immunofluorescence staining was performed as previously described.36 Briefly, B cells were fixed in 4% paraformaldehyde (15 min, room temperature), washed with phosphate-buffered saline, permeabilized (0.1% Triton X-100 in phosphate-buffered saline, 15 min) and attached on precoated poly-L-lysine slides. Slides were blocked in 3% bovine serum albumin, followed by mouse monoclonal anti-NS3 (Abcam; 1:100, ab65407) or anti-core (ThermoFisher Scientific; 1:100, MA1-080) antibody staining. The cells were further incubated with rabbit polyclonal anti-CD19, anti-CD20 (Cell Signaling Technology, Inc., Danvers, MA, USA; 1:400; 3574), anti-CD20 (Abcam; 1:400; ab78237) or anti-B220 antibody (Biolegend; 1:200; #103201). Cells were washed with phosphate-buffered saline and incubated with goat anti-rabbit Alexa Fluor 488 and goat anti-mouse or anti-rat Alexa Fluor 597 (Invitrogen; 1:200). The stained slides were mounted using Vectashield mounting medium (Vector Laboratories Inc., Burlingame, CA, USA). Control slides were similarly processed, except primary antisera were omitted, which yielded no staining. Images were captured using an Olympus FV1000 confocal microscope and processed with Olympus Fluoview Version 1.7c software (Olympus Corporation, Tokyo, Japan).
Co-IP and immunoblotting
Cells were lysed in buffer (20 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 1 mM Na3VO4, 1 mg/ml aprotinin, 1 mg/ml leupterin and 1 mM phenylmethylsulfonyl fluoride) and insoluble material removed by centrifugation. Cell lysates were incubated with protein A- or protein G-sepharose beads precoated with antibodies or control IgG (1 h, 4 °C) and protein complexes washed with lysis buffer (3x, 4 °C). Immunoblotting was performed using standard methods with the following antibodies: anti-NS3 (Abcam, ab18664), anti-pBlk (Tyr388, ab151726), anti-actin (ab8226) anti-pCHK2 (Thr68, Cell Signaling Technology, Inc., CST2661), anti-CARD11 (CST4435), anti-Blk (CST3262), anti-BTK (CST8547), anti-pBTK (Tyr223, CST5082), anti-CD79A (CST3351), anti-CD79A (Tyr182, CST5173), anti-CHK2 (Santa Cruz Biotechnology Inc., Dallas, TX, USA; sc-9064) and anti-SYK (sc-1077).
GST fusion protein pull-down assays
GST fusion proteins were expressed in Escherichia coli BL21(DE3) cells, and purified using glutathione-sepharose beads as previously described.37 Briefly, the GST fusion proteins were coupled to glutathione beads (1 h, 4 °C) and washed with lysis buffer (3x). The immobilized GST fusion proteins were incubated with the cell lysates (1 h, 4 °C). The beads were then washed with lysis buffer (4x) and protein complexes subjected to immunoblotting.
IPs of ribonucleoprotein complexes and microarray data analysis
RIP-Chip was performed as previously described.33 Briefly, cells were lysed in lysis buffer (150 mM KOAc, 2.5 mM Mg(OAc)2, 20 mM K-N-2-hydroxyethylpiperazine-N’-2-ethanesulfonic acid, pH 7.5, dithiothreitol, phenylmethylsulfonyl fluoride, RNasin, and protease inhibitors). Protein lysates (3 mg) were incubated with 100 μl of protein A-sepharose bead suspension, precoated with 30 μg of either mouse anti-HuR antibody or control immunoglobulin IgG (Santa Cruz Biotechnology or BD Biosciences, PharMingen, San Diego, CA, USA, respectively, 1 h, 4 °C). The beads were washed thoroughly with NT2 buffer (50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM MgCl2 and 0.05% Nonidet P-40), treated with 20 units of RNase-free DNase I (15 min, 37 °C), and incubated in 100 μl NT2 buffer containing 0.1% sodium dodecyl sulfate and 0.5 mg/ml proteinase K (20 min, 55 °C). Phenol and chloroform were used to extract RNA from IP material in the presence of GlycoBlue (Ambion, Grand Island, NY, USA).
Microarray data analysis was performed as previously described.32 In short, RNA from IP material was labeled using the Illumina TotalPrep RNA Amplification Kit (Ambion). Human HT-12, version 4.0 gene expression BeadChips containing 47,000 RefSeq transcripts (Illumina Inc., San Diego, CA, USA) were used and raw microarray data were filtered by the detection (P ⩽0.02), normalized by Z-score transformation, and tested for significant differences in signal intensity. The sample quality was analyzed by scatter plot, principal component analysis and gene sample Z-scores based on hierarchical clustering to exclude possible outliers. The genes with larger variances within each comparison group were eliminated according to the analysis of variance test. Genes with fold difference (Z⩾1.5 or ⩽−1.5) were considered to be significantly altered after calculating Z-ratio, false discovery rate, which controls for the expected proportion of false rejected hypothesis (false discovery rate ⩽0.3), and P <0.05. Heatmap was generated using Z-ratio values, representing the top 50 genes with the largest changes in association with HuR in NS3/4A-overexpressing cells relative to control, using ArrayStar (DNAStar, Madison, WA, USA). Genes differentially associated with HuR in studied groups were analyzed by the KEGG pathway map. Ingenuity pathway analysis (http://www.ingenuity.com/; Ingenuity Systems, Redwood City, CA, USA) was used to identify the top network functions in response to NS3/4A overexpression.
Real-time quantitative RT–PCR
RNA from IP material was subjected to reverse transcription using random hexamers and SuperScriptII reverse transcriptase (Invitrogen). The mRNA levels of HuR target were measured by RT–qPCR analysis using iQSYBR Green Supermix (Quanta BioSciences, Inc., Gaithersburg, MD, USA) and a Bio-Rad CFXConnect instrument (Bio-Rad, Hercules, CA, USA). Background binding of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was used as a loading control. Each reaction was carried out in triplicate, and three independent experiments were performed. The primer sequences used were: BCL6, 5'-AACCTGAAAACCCACACTCG-3' and 5'-TTCGCATTTGTAGGGCTTCT-3'; Blk, 5'-TGTGGTCACCAGAGAGCCCATTTA-3' and 5'-TGTCAATCAGCCTTGGAAGGGACA-3'; ALDH1A1, 5'-ACAAGGTGGCCTTCACTGGA-3' and 5'-GCAAACACAATGCAAGGGCT-3'; ALDH1A2, 5'-TGGGTGAGTTTGGCTTACGG-3' and 5'-AGAAACGTGGCAGTCTTGGC-3'; NS3, 5'-CCACATAGACGCCCATTTCT-3' and 5'-GCTCCCAGCCTATACAGCAG-3'; Btk, 5'-TGCAAGGATGTCTGTGAAGC-3' and 5'-GGACAGGCCGAAATCAGATA-3'; CARD11, 5'-TGAATGTAATGCTGGGGACA-3' and 5'-GGCAAGCTGTTCACAAACAA-3'; GAPDH, 5'-CGGAGTCAACGGATTTGGTCGTA-3' and 5'-AGCCTTCTCCATGGTGGTGAAGAC-3'.
Polyribosome fractionation
Cells were incubated for 15 min with 0.1 mg/ml cycloheximide, and then lysed in cytoplasmic lysis buffer containing 150 mM KOAc, 2.5 mM Mg(OAc)2, 20 mM K-N-2-hydroxyethylpiperazine-N’-2-ethanesulfonic acid, pH 7.5, RNaseOUT and protease inhibitor cocktail, and centrifuged for 1 min (13 000 g, 4 °C). Lysates were fractionated through linear sucrose gradients. The eluted fractions were prepared with a fraction collector and their quality was examined at 254 nm using a UV-6 detector (Isco Inc., Lincoln, NE, USA).
Statistical analysis
With the exception of microarray data, which was analyzed as per ‘IPs of ribonucleoprotein complexes and microarray data analysis’ above, data are shown as mean with s.d. from three separate experiments. P-values were calculated using a paired Student's t-test. Statistical significance was determined at P<.05.
References
de Sanjose S, Benavente Y, Vajdic CM, Engels EA, Morton LM, Bracci PM et al. Hepatitis C and non-Hodgkin lymphoma among 4784 cases and 6269 controls from the International Lymphoma Epidemiology Consortium. Clin Gastroenterol Hepatol 2008; 6: 451–458.
Hermine O, Lefrere F, Bronowicki JP, Mariette X, Jondeau K, Eclache-Saudreau V et al. Regression of splenic lymphoma with villous lymphocytes after treatment of hepatitis C virus infection. N Engl J Med 2002; 347: 89–94.
Kasama Y, Sekiguchi S, Saito M, Tanaka K, Satoh M, Kuwahara K et al. Persistent expression of the full genome of hepatitis C virus in B cells induces spontaneous development of B-cell lymphomas in vivo. Blood 2010; 116: 4926–4933.
Beran RK, Pyle AM . Hepatitis C viral NS3-4A protease activity is enhanced by the NS3 helicase. J Biol Chem 2008; 283: 29929–29937.
Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M et al. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci USA 2005; 102: 2992–2997.
Li XD, Sun L, Seth RB, Pineda G, Chen ZJ . Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci USA 2005; 102: 17717–17722.
Lai CK, Jeng KS, Machida K, Cheng YS, Lai MM . Hepatitis C virus NS3/4A protein interacts with ATM, impairs DNA repair and enhances sensitivity to ionizing radiation. Virology 2008; 370: 295–309.
Young RM, Staudt LM . Targeting pathological B cell receptor signalling in lymphoid malignancies. Nat Rev Drug Discov 2013; 12: 229–243.
Davis RE, Ngo VN, Lenz G, Tolar P, Young RM, Romesser PB et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature 2010; 463: 88–92.
Pham TNQ, King D, MacParland SA, McGrath JS, Reddy SB, Bursey FR et al. Hepatitis C virus replicates in the same immune cell subsets in chronic hepatitis C and occult infection. Gastroenterology 2008; 134: 812–822.
Inokuchi M, Ito T, Uchikoshi M, Shimozuma Y, Morikawa K, Nozawa H et al. Infection of B cells with hepatitis C virus for the development of lymphoproliferative disorders in patients with chronic hepatitis C. J Med Virol 2009; 81: 619–627.
Ito M, Murakami K, Suzuki T, Mochida K, Suzuki M, Ikebuchi K et al. Enhanced expression of lymphomagenesis-related genes in peripheral blood B cells of chronic hepatitis C patients. Clin Immunol 2010; 135: 459–465.
Januszkiewicz-Lewandowska D, Wysocki J, Pernak M, Nowicka K, Zawada M, Rembowska J et al. Presence of hepatitis C virus (HCV)-RNA in peripheral blood mononuclear cells in HCV serum negative patients during interferon and ribavirin therapy. Jpn J Infect Dis 2007; 60: 29–32.
Lanford RE, Chavez D, Chisari FV, Sureau C . Lack of detection of negative-strand hepatitis C virus RNA in peripheral blood mononuclear cells and other extrahepatic tissues by the highly strand-specific rTth reverse transcriptase PCR. J Virol 1995; 69: 8079–8083.
Mellor J, Haydon G, Blair C, Livingstone W, Simmonds P . Low level or absent in vivo replication of hepatitis C virus and hepatitis G virus/GB virus C in peripheral blood mononuclear cells. J Gen Virol 1998; 79: 705–714.
Lin L, Fevery J, Hiem Yap S . A novel strand-specific RT-PCR for detection of hepatitis C virus negative-strand RNA (replicative intermediate): evidence of absence or very low level of HCV replication in peripheral blood mononuclear cells. J Virol Methods 2002; 100: 97–105.
Sung VM, Shimodaira S, Doughty AL, Picchio GR, Can H, Yen TS et al. Establishment of B-cell lymphoma cell lines persistently infected with hepatitis C virus in vivo and in vitro: the apoptotic effects of virus infection. J Virol 2003; 77: 2134–2146.
Machida K, Cheng KT, Sung VM, Shimodaira S, Lindsay KL, Levine AM et al. Hepatitis C virus induces a mutator phenotype: enhanced mutations of immunoglobulin and protooncogenes. Proc Natl Acad Sci USA 2004; 101: 4262–4267.
Pham TNQ, MacParland SA, Coffin CS, Lee SS, Bursey FR, Michalak TI . Mitogen-induced upregulation of hepatitis C virus expression in human lymphoid cells. J Gen Virol 2005; 86: 657–666.
Durand T, Di Liberto G, Colman H, Cammas A, Boni S, Marcellin P et al. Occult infection of peripheral B cells by hepatitis C variants which have low translational efficiency in cultured hepatocytes. Gut 2010; 59: 934–942.
Bleesing JJ, Fleisher TA . Human B cells express a CD45 isoform that is similar to murine B220 and is downregulated with acquisition of the memory B-cell marker CD27. Cytometry B Clin Cytom 2003; 51: 1–8.
Bartek J, Falck J, Lukas J . CHK2 kinase–a busy messenger. Nat Rev Mol Cell Biol 2001; 2: 877–886.
Abdelmohsen K, Pullmann Jr R, Lal A, Kim HH, Galban S, Yang X et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol Cell 2007; 25: 543–557.
Mazan-Mamczarz K, Hagner PR, Corl S, Srikantan S, Wood WH, Becker KG et al. Post-transcriptional gene regulation by HuR promotes a more tumorigenic phenotype. Oncogene 2008; 27: 6151–6163.
Okudela K, Woo T, Mitsui H, Suzuki T, Tajiri M, Sakuma Y et al. Downregulation of ALDH1A1 expression in non-small cell lung carcinomas—its clinicopathologic and biological significance. Int J Clin Exp Pathol 2013; 6: 1–12.
Touma SE, Perner S, Rubin MA, Nanus DM, Gudas LJ . Retinoid metabolism and ALDH1A2 (RALDH2) expression are altered in the transgenic adenocarcinoma mouse prostate model. Biochem Pharmacol 2009; 78: 1127–1138.
Pal S, Sullivan DG, Kim S, Lai KK, Kae J, Cotler SJ et al. Productive replication of hepatitis C virus in perihepatic lymph nodes in vivo: implications of HCV lymphotropism. Gastroenterology 2006; 130: 1107–1116.
Ng PP, Kuo CC, Wang S, Einav S, Arcaini L, Paulli M et al. B-cell receptors expressed by lymphomas of hepatitis C virus (HCV)-infected patients rarely react with the viral proteins. Blood 2014; 123: 1512–1515.
Pommier Y, Weinstein JN, Aladjem MI, Kohn KW . Chk2 molecular interaction map and rationale for Chk2 inhibitors. Clin Cancer Res 2006; 12: 2657–2661.
Tan Y, Raychaudhuri P, Costa RH . Chk2 mediates stabilization of the FoxM1 transcription factor to stimulate expression of DNA repair genes. Mol Cell Biol 2007; 27: 1007–1016.
Machida K, McNamara G, Cheng KT, Huang J, Wang CH, Comai L et al. Hepatitis C virus inhibits DNA damage repair through reactive oxygen and nitrogen species and by interfering with the ATM-NBS1/Mre11/Rad50 DNA repair pathway in monocytes and hepatocytes. J Immunol 2010; 185: 6985–6998.
Mazan-Mamczarz K, Hagner PR, Zhang Y, Dai B, Lehrmann E, Becker KG et al. ATM regulates a DNA damage response posttranscriptional RNA operon in lymphocytes. Blood 2011; 117: 2441–2450.
Masuda K, Abdelmohsen K, Kim MM, Srikantan S, Lee EK, Tominaga K et al. Global dissociation of HuR-mRNA complexes promotes cell survival after ionizing radiation. EMBO J 2011; 30: 1040–1053.
Pham TNQ, MacParland SA, Mulrooney PM, Cooksley H, Naoumov NV, Michalak TI . Hepatitis C virus persistence after spontaneous or treatment-induced resolution of hepatitis C. J Virol 2004; 78: 5867–5874.
Sarhan MA, Pham TNQ, Chen AY, Michalak TI . Hepatitis C virus infection of T lymphocytes is mediated by CD5. J Virol 2012; 86: 3723–3735.
Dai B, Zhao XF, Mazan-Mamczarz K, Hagner P, Corl S, Bahassi el M et al. Functional and molecular interactions between ERK and CHK2 in diffuse large B-cell lymphoma. Nat Commun 2011; 2: 402.
Dai B, Zhao XF, Hagner P, Shapiro P, Mazan-Mamczarz K, Zhao S et al. Extracellular signal-regulated kinase positively regulates the oncogenic activity of MCT-1 in diffuse large B-cell lymphoma. Cancer Res 2009; 69: 7835–7843.
Acknowledgements
We are grateful to Dr El Mustapha Bahassi for the gift of CHK2 and its truncated constructs. We thank Dr Michael MC Lai from the Institute of Molecular Biology, Academia Sinica, Taiwan for generously sharing NS3/4A constructs. We are thankful to Dr Michael Gale Jr (University of Washington, WA, USA) for generously sharing a series of FLAG epitope-tagged NS3/4A truncation and deletion mutant plasmids. We are very grateful to Dr Eric Campeau for the use of pLenti-CMVTRE3G and pLenti-CMV-rtTA3-Blast plasmids. We thank Dr Shyamasundaran Kottilil and Dr Patricia M Mulrooney-Cousins for helpful discussions. Flow cytometry analyses were performed at the University of Maryland Marlene and Stewart Greenebaum Cancer Center Flow Cytometry Shared Service. This work was supported in part by a Merit Review Award from the Department of Veterans Affairs (RBG), R01AA017972 (RBG) and R01CA164311 (RBG) from the National Institutes of Health, as well as MOP-126056 (TIM) from the Canadian Institutes of Health Research and the Canada Research Chair Program in Viral Hepatitis/Immunology (TIM). This research was also supported in part by the Intramural Research Program of the NIH, National Institute on Aging (YZ, EL and KGB).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Dai, B., Chen, A., Corkum, C. et al. Hepatitis C virus upregulates B-cell receptor signaling: a novel mechanism for HCV-associated B-cell lymphoproliferative disorders. Oncogene 35, 2979–2990 (2016). https://doi.org/10.1038/onc.2015.364
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.364
This article is cited by
-
Complete regression of an HCV-associated diffuse large B cell lymphoma achieved by chemoimmuno-free antiviral therapy
Annals of Hematology (2021)
-
The clinical impact of hepatitis C virus infection in Egyptian multiple myeloma patients
Journal of the Egyptian National Cancer Institute (2020)
-
Reconstitution of T follicular helper-humoral immune axis with elimination of hepatitis C virus
Scientific Reports (2020)
-
Neutrocyte-to-lymphocyte ratio predicts the presence of a replicative hepatitis C virus strand after therapy with direct-acting antivirals
Clinical and Experimental Medicine (2019)