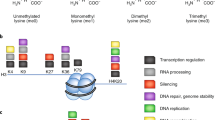
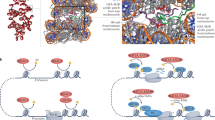
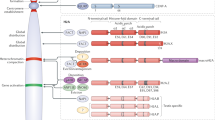
Abstract
Histones undergo several different post-translational modifications that control a variety of physiological processes. These covalent modifications show substantial cross-regulation, providing a wealth of regulatory potential. New insights into the communication between modifications on histones have emerged in recent years. This review assesses the current understanding of cross-regulation of histone modifications and identifies future questions to be addressed in this field.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Kornberg, R.D. Chromatin structure: a repeating unit of histones and DNA. Science 184, 868–871 (1974).
Luger, K., Mader, A.W., Richmond, R.K., Sargent, D.F. & Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 (1997).
Bühler, M. & Moazed, D. Transcription and RNA interference in heterochromatic gene silencing. Nat. Struct. Mol. Biol. 14, 1008–1016 (2007).
Li, B., Carey, M. & Workman, J.L. The role of chromatin during transcription. Cell 128, 707–719 (2007).
Groth, A., Rocha, W., Verreault, A. & Almouzni, G. Chromatin challenges during DNA replication and repair. Cell 128, 721–733 (2007).
Allfrey, V.G., Faulkner, R. & Mirsky, A.E. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. USA 51, 786–794 (1964).
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007).
Strahl, B.D. & Allis, C.D. The language of covalent histone modifications. Nature 403, 41–45 (2000).
Turner, B.M. Histone acetylation and an epigenetic code. Bioessays 22, 836–845 (2000).
Hyland, E.M. et al. Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae. Mol. Cell. Biol. 25, 10060–10070 (2005).
Turner, B.M. Reading signals on the nucleosome with a new nomenclature for modified histones. Nat. Struct. Mol. Biol. 12, 110–112 (2005).
Cuthbert, G.L. et al. Histone deimination antagonizes arginine methylation. Cell 118, 545–553 (2004).
Nakayama, J., Rice, J.C., Strahl, B.D., Allis, C.D. & Grewal, S.I. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292, 110–113 (2001).
Rea, S. et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599 (2000).
Hassan, Y.I. & Zempleni, J. Epigenetic regulation of chromatin structure and gene function by biotin. J. Nutr. 136, 1763–1765 (2006).
Camporeale, G., Oommen, A.M., Griffin, J.B., Sarath, G. & Zempleni, J. K12-biotinylated histone H4 marks heterochromatin in human lymphoblastoma cells. J. Nutr. Biochem. published online, doi:10.1016/j.jnutbio.2006.12.014 (13 April 2007).
Nathan, D. et al. Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications. Genes Dev. 20, 966–976 (2006).
Morris, S.A. et al. Identification of histone H3 lysine 36 acetylation as a highly conserved histone modification. J. Biol. Chem. 282, 7632–7640 (2007).
Aihara, H. et al. Nucleosomal histone kinase-1 phosphorylates H2A Thr 119 during mitosis in the early Drosophila embryo. Genes Dev. 18, 877–888 (2004).
Ivanovska, I., Khandan, T., Ito, T. & Orr-Weaver, T.L. A histone code in meiosis: the histone kinase, NHK-1, is required for proper chromosomal architecture in Drosophila oocytes. Genes Dev. 19, 2571–2582 (2005).
Shiio, Y. & Eisenman, R.N. Histone sumoylation is associated with transcriptional repression. Proc. Natl. Acad. Sci. USA 100, 13225–13230 (2003).
Shin, J.A. et al. SUMO modification is involved in the maintenance of heterochromatin stability in fission yeast. Mol. Cell 19, 817–828 (2005).
Suka, N., Suka, Y., Carmen, A.A., Wu, J. & Grunstein, M. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell 8, 473–479 (2001).
Ahn, S.H., Diaz, R.L., Grunstein, M. & Allis, C.D. Histone H2B deacetylation at lysine 11 is required for yeast apoptosis induced by phosphorylation of H2B at serine 10. Mol. Cell 24, 211–220 (2006).
Cheung, W.L. et al. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell 113, 507–517 (2003).
Thorne, A.W., Kmiciek, D., Mitchelson, K., Sautiere, P. & Crane-Robinson, C. Patterns of histone acetylation. Eur. J. Biochem. 193, 701–713 (1990).
Shilatifard, A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 75, 243–269 (2006).
Shahbazian, M.D., Zhang, K. & Grunstein, M. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol. Cell 19, 271–277 (2005).
Dehe, P.M. et al. Histone H3 lysine 4 mono-methylation does not require ubiquitination of histone H2B. J. Mol. Biol. 353, 477–484 (2005).
Henry, K.W. et al. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 17, 2648–2663 (2003).
Nelson, C.J., Santos-Rosa, H. & Kouzarides, T. Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell 126, 905–916 (2006).
Hsu, J.Y. et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102, 279–291 (2000).
Cheung, P. et al. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell 5, 905–915 (2000).
Lo, W.S. et al. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell 5, 917–926 (2000).
Edmondson, D.G. et al. Site-specific loss of acetylation upon phosphorylation of histone H3. J. Biol. Chem. 277, 29496–29502 (2002).
Lee, D.Y., Northrop, J.P., Kuo, M.H. & Stallcup, M.R. Histone H3 lysine 9 methyltransferase G9a is a transcriptional coactivator for nuclear receptors. J. Biol. Chem. 281, 8476–8485 (2006).
Fischle, W. et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438, 1116–1122 (2005).
Lachner, M., O'Carroll, D., Rea, S., Mechtler, K. & Jenuwein, T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120 (2001).
Bannister, A.J. et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124 (2001).
Hirota, T., Lipp, J.J., Toh, B.H. & Peters, J.M. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature 438, 1176–1180 (2005).
Mateescu, B., England, P., Halgand, F., Yaniv, M. & Muchardt, C. Tethering of HP1 proteins to chromatin is relieved by phosphoacetylation of histone H3. EMBO Rep. 5, 490–496 (2004).
Daujat, S., Zeissler, U., Waldmann, T., Happel, N. & Schneider, R. HP1 binds specifically to Lys26-methylated histone H1.4, whereas simultaneous Ser27 phosphorylation blocks HP1 binding. J. Biol. Chem. 280, 38090–38095 (2005).
Czermin, B. et al. Physical and functional association of SU(VAR)3–9 and HDAC1 in Drosophila. EMBO Rep. 2, 915–919 (2001).
Vaute, O., Nicolas, E., Vandel, L. & Trouche, D. Functional and physical interaction between the histone methyl transferase Suv39H1 and histone deacetylases. Nucleic Acids Res. 30, 475–481 (2002).
Zhang, C.L., McKinsey, T.A. & Olson, E.N. Association of class II histone deacetylases with heterochromatin protein 1: potential role for histone methylation in control of muscle differentiation. Mol. Cell. Biol. 22, 7302–7312 (2002).
Wang, H. et al. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol. Cell 8, 1207–1217 (2001).
Thomas, C.E., Kelleher, N.L. & Mizzen, C.A. Mass spectrometric characterization of human histone H3: a bird's eye view. J. Proteome Res. 5, 240–247 (2006).
Johnson, L. et al. Mass spectrometry analysis of Arabidopsis histone H3 reveals distinct combinations of post-translational modifications. Nucleic Acids Res. 32, 6511–6518 (2004).
Zhang, K. et al. Differentiation between peptides containing acetylated or tri-methylated lysines by mass spectrometry: an application for determining lysine 9 acetylation and methylation of histone H3. Proteomics 4, 1–10 (2004).
Taverna, S.D., Li, H., Ruthenburg, A.J., Allis, C.D. & Patel, D.J. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 14, 1025–1040 (2007).
Vakoc, C.R., Mandat, S.A., Olenchock, B.A. & Blobel, G.A. Histone H3 lysine 9 methylation and HP1γ are associated with transcription elongation through mammalian chromatin. Mol. Cell 19, 381–391 (2005).
Nishioka, K. et al. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 16, 479–489 (2002).
Nightingale, K.P. et al. Cross-talk between histone modifications in response to histone deacetylase inhibitors: MLL4 links histone H3 acetylation and histone H3K4 methylation. J. Biol. Chem. 282, 4408–4416 (2007).
Martin, D.G., Grimes, D.E., Baetz, K. & Howe, L. Methylation of histone H3 mediates the association of the NuA3 histone acetyltransferase with chromatin. Mol. Cell. Biol. 26, 3018–3028 (2006).
Martin, D.G. et al. The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol. Cell. Biol. 26, 7871–7879 (2006).
Taverna, S.D. et al. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol. Cell 24, 785–796 (2006).
Morillon, A., Karabetsou, N., Nair, A. & Mellor, J. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol. Cell 18, 723–734 (2005).
Govind, C.K., Zhang, F., Qiu, H., Hofmeyer, K. & Hinnebusch, A.G. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol. Cell 25, 31–42 (2007).
Milne, T.A. et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell 10, 1107–1117 (2002).
Pray-Grant, M.G., Daniel, J.A., Schieltz, D., Yates, J.R. III & Grant, P.A. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature 433, 434–438 (2005).
Sims, R.J. III et al. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J. Biol. Chem. 280, 41789–41792 (2005).
Dou, Y. et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell 121, 873–885 (2005).
Shi, X. et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442, 96–99 (2006).
Hazzalin, C.A. & Mahadevan, L.C. Dynamic acetylation of all lysine 4-methylated histone H3 in the mouse nucleus: analysis at c-fos and c-jun. PLoS Biol. 3, e393 (2005).
Lee, M.G., Norman, J., Shilatifard, A. & Shiekhattar, R. Physical and functional association of a trimethyl H3K4 demethylase and Ring6a/MBLR, a polycomb-like protein. Cell 128, 877–887 (2007).
Schuettengruber, B., Chourrout, D., Vervoort, M., Leblanc, B. & Cavalli, G. Genome regulation by polycomb and trithorax proteins. Cell 128, 735–745 (2007).
Lee, M.G., Wynder, C., Cooch, N. & Shiekhattar, R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 437, 432–435 (2005).
Shi, Y.J. et al. Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell 19, 857–864 (2005).
Lee, M.G. et al. Functional interplay between histone demethylase and deacetylase enzymes. Mol. Cell. Biol. 26, 6395–6402 (2006).
Joshi, A.A. & Struhl, K. Eaf3 chromodomain interaction with methylated H3–K36 links histone deacetylation to Pol II elongation. Mol. Cell 20, 971–978 (2005).
Keogh, M.C. et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123, 593–605 (2005).
Carrozza, M.J. et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123, 581–592 (2005).
Brown, M.A., Sims, R.J. III, Gottlieb, P.D. & Tucker, P.W. Identification and characterization of Smyd2: a split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex. Mol. Cancer 5, 26 (2006).
Tompa, R. & Madhani, H.D. Histone H3 lysine 36 methylation antagonizes silencing in Saccharomyces cerevisiae independently of the Rpd3S histone deacetylase complex. Genetics 175, 585–593 (2007).
Sims, J.K., Houston, S.I., Magazinnik, T. & Rice, J.C. A trans-tail histone code defined by monomethylated H4 Lys-20 and H3 Lys-9 demarcates distinct regions of silent chromatin. J. Biol. Chem. 281, 12760–12766 (2006).
Yin, Y. et al. SET8 recognizes the sequence RHRK20VLRDN within the N terminus of histone H4 and mono-methylates lysine 20. J. Biol. Chem. 280, 30025–30031 (2005).
Fingerman, I.M., Li, H.C. & Briggs, S.D. A charge-based interaction between histone H4 and Dot1 is required for H3K79 methylation and telomere silencing: identification of a new trans-histone pathway. Genes Dev. 21, 2018–2029 (2007).
Nishioka, K. et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol. Cell 9, 1201–1213 (2002).
Kourmouli, N. et al. Heterochromatin and tri-methylated lysine 20 of histone H4 in animals. J. Cell Sci. 117, 2491–2501 (2004).
Sarg, B., Helliger, W., Talasz, H., Koutzamani, E. & Lindner, H.H. Histone H4 hyperacetylation precludes histone H4 lysine 20 trimethylation. J. Biol. Chem. 279, 53458–53464 (2004).
Talasz, H., Lindner, H.H., Sarg, B. & Helliger, W. Histone H4-lysine 20 monomethylation is increased in promoter and coding regions of active genes and correlates with hyperacetylation. J. Biol. Chem. 280, 38814–38822 (2005).
Wang, H. et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 293, 853–857 (2001).
Strahl, B.D. et al. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr. Biol. 11, 996–1000 (2001).
Yu, M.C., Lamming, D.W., Eskin, J.A., Sinclair, D.A. & Silver, P.A. The role of protein arginine methylation in the formation of silent chromatin. Genes Dev. 20, 3249–3254 (2006).
Fabbrizio, E. et al. Negative regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO Rep. 3, 641–645 (2002).
Pal, S. et al. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol. Cell. Biol. 23, 7475–7487 (2003).
Le Guezennec, X. et al. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol. Cell. Biol. 26, 843–851 (2006).
Zhang, K. et al. Histone acetylation and deacetylation: identification of acetylation and methylation sites of HeLa histone H4 by mass spectrometry. Mol. Cell. Proteomics 1, 500–508 (2002).
Utley, R.T., Lacoste, N., Jobin-Robitaille, O., Allard, S. & Cote, J. Regulation of NuA4 histone acetyltransferase activity in transcription and DNA repair by phosphorylation of histone H4. Mol. Cell. Biol. 25, 8179–8190 (2005).
Bode, A.M. & Dong, Z. Post-translational modification of p53 in tumorigenesis. Nat. Rev. Cancer 4, 793–805 (2004).
Xirodimas, D.P., Saville, M.K., Bourdon, J.C., Hay, R.T. & Lane, D.P. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell 118, 83–97 (2004).
Huang, J. et al. Repression of p53 activity by Smyd2-mediated methylation. Nature 444, 629–632 (2006).
Youmell, M., Park, S.J., Basu, S. & Price, B.D. Regulation of the p53 protein by protein kinase C alpha and protein kinase C zeta. Biochem. Biophys. Res. Commun. 245, 514–518 (1998).
Chuikov, S. et al. Regulation of p53 activity through lysine methylation. Nature 432, 353–360 (2004).
Liu, L. et al. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 19, 1202–1209 (1999).
Zhang, K. et al. The Set1 methyltransferase opposes Ipl1 aurora kinase functions in chromosome segregation. Cell 122, 723–734 (2005).
Cheeseman, I.M. et al. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 111, 163–172 (2002).
Grégoire, S. et al. Control of MEF2 transcriptional activity by coordinated phosphorylation and sumoylation. J. Biol. Chem. 281, 4423–4433 (2006).
Grégoire, S. & Yang, X. Association with class IIa histone deacetylases upregulates the sumoylation of MEF2 transcription factors. Mol. Cell Biol. 25, 2273–2287 (2005).
Shalizi, A. et al. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science 311, 1012–1017 (2006).
Moore, J.D., Yazgan, O., Ataian, Y. & Krebs, J.E. Diverse roles for histone H2A modifications in DNA damage response pathways in yeast. Genetics 176, 15–25 (2006).
Taverna, S.D. et al. Long-distance combinatorial linkage between methylation and acetylation on histone H3 N termini. Proc. Natl. Acad. Sci. USA 104, 2086–2091 (2007).
Bernstein, B.E. et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 (2006).
Acknowledgements
We thank A. Loyola and R. Chosed for critical comments and suggestions. J.A.L. is supported in part by a fellowship by the American Legion Auxiliary. Aspects of this work were supported by US National Institutes of Health grant GM067718 and a grant from the Robert A. Welch Foundation (G1371) to S.Y.R.D. We apologize to colleagues whose work was not cited owing to space limitations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Latham, J., Dent, S. Cross-regulation of histone modifications. Nat Struct Mol Biol 14, 1017–1024 (2007). https://doi.org/10.1038/nsmb1307
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb1307
This article is cited by
-
Mechanisms of gene regulation by histone degradation in adaptation of yeast: an overview of recent advances
Archives of Microbiology (2022)
-
LATS1 K751 acetylation blocks activation of Hippo signalling and switches LATS1 from a tumor suppressor to an oncoprotein
Science China Life Sciences (2022)
-
Effects of acute heat stress on protein expression and histone modification in the adrenal gland of male layer-type country chickens
Scientific Reports (2021)
-
Transgenerational effect of drug-mediated inhibition of LSD1 on eye pigment expression in Drosophila
BMC Ecology (2020)
-
UHRF1-KAT7-mediated regulation of TUSC3 expression via histone methylation/acetylation is critical for the proliferation of colon cancer cells
Oncogene (2020)