Abstract
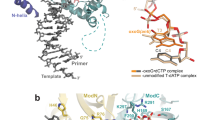
The aromatic amine carcinogen 2-aminofluorene (AF) forms covalent adducts with DNA, predominantly with guanine at the C8 position. Such lesions are bypassed by Y-family polymerases such as Dpo4 via error-free and error-prone mechanisms. We show that Dpo4 catalyzes elongation from a correct 3′-terminal cytosine opposite [AF]G in a nonrepetitive template sequence with low efficiency. This extension leads to cognate full-length product, as well as mis-elongated products containing base mutations and deletions. Crystal structures of the Dpo4 ternary complex, with the 3′-terminal primer cytosine base opposite [AF]G in the anti conformation and with the AF moiety positioned in the major groove, reveal both accurate and misalignment-mediated mutagenic extension pathways. The mutagenic template–primer–dNTP arrangement is promoted by interactions between the polymerase and the bulky lesion rather than by a base pair–stabilized misaligment. Further extension leads to semitargeted mutations via this proposed polymerase-guided mechanism.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Kadlubar, F.F. DNA adducts of carcinogenic aromatic amines. IARC Sci. Publ. 125, 199–216 (1994).
Turesky, R.J. Formation and biochemistry of carcinogenic heterocyclic aromatic amines in cooked meats. Toxicol. Lett. 168, 219–227 (2007).
Clapp, R.W., Jacobs, M.M. & Loechler, E.L. Environmental and occupational causes of cancer: new evidence 2005-2007. Rev. Environ. Health 23, 1–37 (2008).
Heflich, R.H. & Neft, R.E. Genetic toxicity of 2-acetylaminofluorene, 2-aminofluorene and some of their metabolites and model metabolites. Mutat. Res. 318, 73–114 (1994).
Shibutani, S., Suzuki, N., Tan, X., Johnson, F. & Grollman, A.P. Influence of flanking sequence context on the mutagenicity of acetylaminofluorene-derived DNA adducts in mammalian cells. Biochemistry 40, 3717–3722 (2001).
Watt, D.L., Utzat, C.D., Hilario, P. & Basu, A.K. Mutagenicity of the 1-nitropyrene-DNA adduct N-(deoxyguanosin-8-yl)-1-aminopyrene in mammalian cells. Chem. Res. Toxicol. 20, 1658–1664 (2007).
Tan, X., Suzuki, N., Grollman, A.P. & Shibutani, S. Mutagenic events in Escherichia coli and mammalian cells generated in response to acetylaminofluorene-derived DNA adducts positioned in the Nar I restriction enzyme site. Biochemistry 41, 14255–14262 (2002).
Mozzherin, D.J., Shibutani, S., Tan, C.K., Downey, K.M. & Fisher, P.A. Proliferating cell nuclear antigen promotes DNA synthesis past template lesions by mammalian DNA polymerase delta. Proc. Natl. Acad. Sci. USA 94, 6126–6131 (1997).
Doisy, R. & Tang, M.S. Effect of aminofluorene and (acetylamino)fluorene adducts on the DNA replication mediated by Escherichia coli polymerases I (Klenow fragment) and III. Biochemistry 34, 4358–4368 (1995).
Lindsley, J.E. & Fuchs, R.P. Use of single-turnover kinetics to study bulky adduct bypass by T7 DNA polymerase. Biochemistry 33, 764–772 (1994).
Belguise-Valladier, P. & Fuchs, R.P. N-2-aminofluorene and N-2 acetylaminofluorene adducts: the local sequence context of an adduct and its chemical structure determine its replication properties. J. Mol. Biol. 249, 903–913 (1995).
Miller, H. & Grollman, A.P. Kinetics of DNA polymerase I (Klenow fragment exo-) activity on damaged DNA templates: effect of proximal and distal template damage on DNA synthesis. Biochemistry 36, 15336–15342 (1997).
Shibutani, S. & Grollman, A.P. On the mechanism of frameshift (deletion) mutagenesis in vitro. J. Biol. Chem. 268, 11703–11710 (1993).
Lone, S. & Romano, L.J. The role of specific amino acid residues in the active site of Escherichia coli DNA polymerase I on translesion DNA synthesis across from and past an N-2-aminofluorene adduct. Biochemistry 46, 2599–2607 (2007).
Yang, W. & Woodgate, R. What a difference a decade makes: insights into translesion DNA synthesis. Proc. Natl. Acad. Sci. USA 104, 15591–15598 (2007).
McCulloch, S.D. & Kunkel, T.A. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 18, 148–161 (2008).
Waters, L.S. et al. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol. Mol. Biol. Rev. 73, 134–154 (2009).
Koffel-Schwartz, N., Coin, F., Veaute, X. & Fuchs, R.P. Cellular strategies for accommodating replication-hindering adducts in DNA: control by the SOS response in Escherichia coli. Proc. Natl. Acad. Sci. USA 93, 7805–7810 (1996).
Tebbs, R.S. & Romano, L.J. Mutagenesis at a site-specifically modified NarI sequence by acetylated and deacetylated aminofluorene adducts. Biochemistry 33, 8998–9006 (1994).
Reifferscheid, G. & Heil, J. Validation of the SOS/umu test using test results of 486 chemicals and comparison with the Ames test and carcinogenicity data. Mutat. Res. 369, 129–145 (1996).
Suzuki, N. et al. Translesional synthesis past acetylaminofluorene-derived DNA adducts catalyzed by human DNA polymerase κ and Escherichia coli DNA polymerase IV. Biochemistry 40, 15176–15183 (2001).
Vooradi, V. & Romano, L.J. Effect of N-2-acetylaminofluorene and 2-aminofluorene adducts on DNA binding and synthesis by yeast DNA polymerase β. Biochemistry 48, 4209–4216 (2009).
Steitz, T.A. & Yin, Y.W. Accuracy, lesion bypass, strand displacement and translocation by DNA polymerases. Phil. Trans. R. Soc. Lond. B 359, 17–23 (2004).
Johnson, S.J. & Beese, L.S. Structures of mismatch replication errors observed in a DNA polymerase. Cell 116, 803–816 (2004).
Swan, M.K., Johnson, R.E., Prakash, L., Prakash, S. & Aggarwal, A.K. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase δ. Nat. Struct. Mol. Biol. 16, 979–986 (2009).
Lone, S. et al. Human DNA polymerase kappa encircles DNA: implications for mismatch extension and lesion bypass. Mol. Cell 25, 601–614 (2007).
Alt, A. et al. Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase eta. Science 318, 967–970 (2007).
Mizukami, S., Kim, T.W., Helquist, S.A. & Kool, E.T. Varying DNA base-pair size in subangstrom increments: evidence for a loose, not large, active site in low-fidelity Dpo4 polymerase. Biochemistry 45, 2772–2778 (2006).
Broyde, S., Wang, L., Rechkoblit, O., Geacintov, N.E. & Patel, D.J. Lesion processing: high-fidelity versus lesion-bypass DNA polymerases. Trends Biochem. Sci. 33, 209–219 (2008).
Hsu, G.W. et al. Observing translesion synthesis of an aromatic amine DNA adduct by a high-fidelity DNA polymerase. J. Biol. Chem. 279, 50280–50285 (2004).
Dutta, S. et al. Crystal structures of 2-acetylaminofluorene and 2-aminofluorene in complex with T7 DNA polymerase reveal mechanisms of mutagenesis. Proc. Natl. Acad. Sci. USA 101, 16186–16191 (2004).
Burnouf, D.Y. & Wagner, J.E. Kinetics of deoxy-CTP incorporation opposite a dG-C8-N-2-aminofluorene adduct by a high-fidelity DNA polymerase. J. Mol. Biol. 386, 951–961 (2009).
Patel, D.J. et al. Nuclear magnetic resonance solution structures of covalent aromatic amine-DNA adducts and their mutagenic relevance. Chem. Res. Toxicol. 11, 391–407 (1998).
Meneni, S.R. et al. Spectroscopic and theoretical insights into sequence effects of aminofluorene-induced conformational heterogeneity and nucleotide excision repair. Biochemistry 46, 11263–11278 (2007).
Norman, D. et al. NMR and computational characterization of the N-(deoxyguanosin-8-yl)aminofluorene adduct [(AF)G] opposite adenosine in DNA: (AF)G[syn].A[anti] pair formation and its pH dependence. Biochemistry 28, 7462–7476 (1989).
Jain, N., Meneni, S., Jain, V. & Cho, B.P. Influence of flanking sequence context on the conformational flexibility of aminofluorene-modified dG adduct in dA mismatch DNA duplexes. Nucleic Acids Res. 37, 1628–1637 (2009).
Kunkel, T.A. Misalignment-mediated DNA synthesis errors. Biochemistry 29, 8003–8011 (1990).
Ripley, L.S. Frameshift mutation: determinants of specificity. Annu. Rev. Genet. 24, 189–213 (1990).
Tippin, B., Kobayashi, S., Bertram, J.G. & Goodman, M.F. To slip or skip, visualizing frameshift mutation dynamics for error-prone DNA polymerases. J. Biol. Chem. 279, 45360–45368 (2004).
Lambert, I.B., Napolitano, R.L. & Fuchs, R.P. Carcinogen-induced frameshift mutagenesis in repetitive sequences. Proc. Natl. Acad. Sci. USA 89, 1310–1314 (1992).
Shibutani, S., Suzuki, N. & Grollman, A.P. Mechanism of frameshift (deletion) generated by acetylaminofluorene-derived DNA adducts in vitro. Biochemistry 43, 15929–15935 (2004).
Stover, J.S., Chowdhury, G., Zang, H., Guengerich, F.P. & Rizzo, C.J. Translesion synthesis past the C8- and N2-deoxyguanosine adducts of the dietary mutagen 2-Amino-3-methylimidazo[4,5-f]quinoline in the NarI recognition sequence by prokaryotic DNA polymerases. Chem. Res. Toxicol. 19, 1506–1517 (2006).
Bacolod, M.D., Krishnasamy, R. & Basu, A.K. Mutagenicity of the 1-nitropyrene-DNA adduct N-(deoxyguanosin-8-yl)-1-aminopyrene in Escherichia coli located in a nonrepetitive CGC sequence. Chem. Res. Toxicol. 13, 523–528 (2000).
Garcia-Diaz, M., Bebenek, K., Krahn, J.M., Pedersen, L.C. & Kunkel, T.A. Structural analysis of strand misalignment during DNA synthesis by a human DNA polymerase. Cell 124, 331–342 (2006).
Wilson, R.C. & Pata, J.D. Structural insights into the generation of single-base deletions by the Y family DNA polymerase dbh. Mol. Cell 29, 767–779 (2008).
Bauer, J. et al. A structural gap in Dpo4 supports mutagenic bypass of a major benzo[a]pyrene dG adduct in DNA through template misalignment. Proc. Natl. Acad. Sci. USA 104, 14905–14910 (2007).
Wang, F. & Yang, W. Structural insight into translesion synthesis by DNA Pol II. Cell 139, 1279–1289 (2009).
Ling, H., Boudsocq, F., Woodgate, R. & Yang, W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell 107, 91–102 (2001).
Rechkoblit, O. et al. Stepwise translocation of Dpo4 polymerase during error-free bypass of an oxoG lesion. PLoS Biol. 4, e11 (2006).
Vaisman, A., Ling, H., Woodgate, R. & Yang, W. Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis. EMBO J. 24, 2957–2967 (2005).
Hunter, W.N., Brown, T. & Kennard, O. Structural features and hydration of d(C-G-C-G-A-A-T-T-A-G-C-G); a double helix containing two G.A mispairs. J. Biomol. Struct. Dyn. 4, 173–191 (1986).
Colis, L.C., Raychaudhury, P. & Basu, A.K. Mutational specificity of gamma-radiation-induced guanine-thymine and thymine-guanine intrastrand cross-links in mammalian cells and translesion synthesis past the guanine-thymine lesion by human DNA polymerase eta. Biochemistry 47, 8070–8079 (2008).
Hendel, A., Ziv, O., Gueranger, Q., Geacintov, N. & Livneh, Z. Reduced efficiency and increased mutagenicity of translesion DNA synthesis across a TT cyclobutane pyrimidine dimer, but not a TT 6-4 photoproduct, in human cells lacking DNA polymerase eta. DNA Repair (Amst.) 7, 1636–1646 (2008).
Bailey, E.A., Iyer, R.S., Stone, M.P., Harris, T.M. & Essigmann, J.M. Mutational properties of the primary aflatoxin B1-DNA adduct. Proc. Natl. Acad. Sci. USA 93, 1535–1539 (1996).
Bishop, R.E., Pauly, G.T. & Moschel, R.C. O6-ethylguanine and O6-benzylguanine incorporated site-specifically in codon 12 of the rat H-ras gene induce semi-targeted as well as targeted mutations in Rat4 cells. Carcinogenesis 17, 849–856 (1996).
Jelinsky, S.A., Liu, T., Geacintov, N.E. & Loechler, E.L. The major, N2-Gua adduct of the (+)-anti-benzo[a]pyrene diol epoxide is capable of inducing G-->A and G-->C, in addition to G-->T, mutations. Biochemistry 34, 13545–13553 (1995).
Xu, P., Oum, L., Lee, Y.C., Geacintov, N.E. & Broyde, S. Visualizing sequence-governed nucleotide selectivities and mutagenic consequences through a replicative cycle: processing of a bulky carcinogen N(2)-dG lesion in a Y-family DNA polymerase. Biochemistry 48, 4677–4690 (2009).
Bresson, A. & Fuchs, R.P. Lesion bypass in yeast cells: Pol β participates in a multi-DNA polymerase process. EMBO J. 21, 3881–3887 (2002).
Eckel, L.M. & Krugh, T.R. 2-Aminofluorene modified DNA duplex exists in two interchangeable conformations. Nat. Struct. Biol. 1, 89–94 (1994).
Nair, D.T., Johnson, R.E., Prakash, L., Prakash, S. & Aggarwal, A.K. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science 309, 2219–2222 (2005).
Acknowledgements
We thank Y.Cheng for help with expression and purification of Dpo4, A. Serganov for data collection from the [AF]G·C-1 and [AF]G·A-2 crystals and L. Wang (New York Univ.) for providing the coordinates of the [AF]G-modified DNA duplexes. The research was supported by US National Institutes of Health grants CA46533 to D.J.P., CA75449 to S.B. and CA99194 to N.E.G. Partial support for computational infrastructure and computer systems management was also provided to S.B. by CA28038. We would like to thank the staff at the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source (APS), Argonne National Laboratory, supported by award RR-15301 from the National Center for Research Resources at the National Institute of Health, for assistance with data collection. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
Author information
Authors and Affiliations
Contributions
O.R. conducted and interpreted all the structural and kinetic measurements under the supervision of D.J.P., S.B. and N.E.G.; A.K. was responsible for the synthesis and purification of [AF]G-containing DNAs under the supervision of N.E.G.; L.M. was involved in specific aspects of crystal structure refinement. The paper was written by O.R., D.J.P., S.B. and N.E.G.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10, Supplementary Table 1 and Supplementary Data (PDF 771 kb)
Rights and permissions
About this article
Cite this article
Rechkoblit, O., Kolbanovskiy, A., Malinina, L. et al. Mechanism of error-free and semitargeted mutagenic bypass of an aromatic amine lesion by Y-family polymerase Dpo4. Nat Struct Mol Biol 17, 379–388 (2010). https://doi.org/10.1038/nsmb.1771
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1771