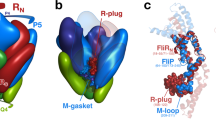
Abstract
R21, the lysozyme of coliphage 21, has an N-terminal signal-anchor-release (SAR) domain that directs its secretion in a membrane-tethered, inactive form and then its release and activation in the periplasm. Both genetic and crystallographic studies show that the SAR domain, once extracted from the bilayer, refolds into the body of the enzyme and effects muralytic activation by repositioning one residue of the canonical lysozyme catalytic triad.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Xu, M., Struck, D.K., Deaton, J., Wang, I.N. & Young, R. Proc. Natl. Acad. Sci. USA 101, 6415–6420 (2004).
Xu, M. et al. Science 307, 113–117 (2005).
Park, T., Struck, D.K., Deaton, J.F. & Young, R. Proc. Natl. Acad. Sci. USA 103, 19713–19718 (2006).
Park, T., Struck, D.K., Dankenbring, C.A. & Young, R. J. Bacteriol. 189, 9135–9139 (2007).
Lei, S.P., Lin, H.C., Wang, S.S., Callaway, J. & Wilcox, G. J. Bacteriol. 169, 4379–4383 (1987).
Mooers, B.H. & Matthews, B.W. Acta Crystallogr. Sect D Biol. Crystallogr. 62, 165–176 (2006).
Bell, J.A. et al. Proteins 10, 10–21 (1991).
Strynadka, N.C. & James, M.N. EXS 75, 185–222 (1996).
Rennell, D., Bouvier, S.E., Hardy, L.W. & Poteete, A.R. J. Mol. Biol. 222, 67–88 (1991).
Hardy, L.W. & Poteete, A.R. Biochemistry 30, 9457–9463 (1991).
Asakura, M. et al. Proc. Natl. Acad. Sci. USA 104, 14483–14488 (2007).
Plunkett, G. III, Rose, D.J., Durfee, T.J. & Blattner, F.R. J. Bacteriol. 181, 1767–1778 (1999).
O'Brien, A.D., Marques, L.R., Kerry, C.F., Newland, J.W. & Holmes, R.K. Microb. Pathog. 6, 381–390 (1989).
Ochoa, T.J., Chen, J., Walker, C.M., Gonzales, E. & Cleary, T.G. Antimicrob. Agents Chemother. 51, 2837–2841 (2007).
Acknowledgements
We would like to acknowledge the contribution of E. McKee and E. Caronna in the structure determination of aR21. The work of Q.S., A.A. and J.C.S. was supported by funds to J.C.S. from the Robert A. Welch Foundation A-0015 and by US National Institutes of Health grant PO1AIO60342. The work of G.F.K., M.X. and R.Y. was supported by National Institutes of Health grant NIGMS27099 and by the Program for Membrane Structure and Function, a Program of Excellence grant from the Office of the Vice President for Research at Texas A&M University. Use of the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under contract no. DE-AC02-06CH11357.
Author information
Authors and Affiliations
Contributions
Q.S. determined the structure of iR21 and refined the structure of aR21 initially crystallized by A.A. Q.S. also performed the in vitro assays. G.F.K. did the molecular biology, genetics, physiology and bioinformatics. M.X. provided key constructs. R.Y. and J.C.S. provided supervision. The writing of the manuscript was a collaborative effort by Q.S., G.F.K., J.C.S. and R.Y.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1 and 2 and Supplementary Figures 1–5 (PDF 700 kb)
Rights and permissions
About this article
Cite this article
Sun, Q., Kuty, G., Arockiasamy, A. et al. Regulation of a muralytic enzyme by dynamic membrane topology. Nat Struct Mol Biol 16, 1192–1194 (2009). https://doi.org/10.1038/nsmb.1681
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1681
This article is cited by
-
Phage spanins: diversity, topological dynamics and gene convergence
BMC Bioinformatics (2018)
-
Modular endolysin of Burkholderia AP3 phage has the largest lysozyme-like catalytic subunit discovered to date and no catalytic aspartate residue
Scientific Reports (2017)
-
Phage lysis: Three steps, three choices, one outcome
Journal of Microbiology (2014)
-
The Caulobacter crescentus phage phiCbK: genomics of a canonical phage
BMC Genomics (2012)