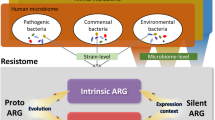
Abstract
Metagenomic studies have shown that antibiotic resistance genes are ubiquitous in the environment, which has led to the suggestion that there is a high risk that these genes will spread to bacteria that cause human infections. If this is true, estimating the real risk of dissemination of resistance genes from environmental reservoirs to human pathogens is therefore very difficult. In this Opinion article, we analyse the current definitions of antibiotic resistance and antibiotic resistance genes, and we describe the bottlenecks that affect the transfer of antibiotic resistance genes to human pathogens. We propose rules for estimating the risks associated with genes that are present in environmental resistomes by evaluating the likelihood of their introduction into human pathogens, and the consequences of such events for the treatment of infections.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
World Health Organization. Antimicrobial Resistance: Global Report on Surveillance. (WHO Press, 2014).
Kesselheim, A. S. & Outterson, K. Fighting antibiotic resistance: marrying new financial incentives to meeting public health goals. Health Aff. (Millwood) 29, 1689–1696 (2010).
Baquero, F. Metagenomic epidemiology: a public health need for the control of antimicrobial resistance. Clin. Microbiol. Infect. 18 (Suppl. 4), 67–73 (2012).
Allen, H. K. et al. Call of the wild: antibiotic resistance genes in natural environments. Nature Rev. Microbiol. 8, 251–259 (2010).
Forsberg, K. J. et al. The shared antibiotic resistome of soil bacteria and human pathogens. Science 337, 1107–1111 (2012).
Sommer, M. O., Dantas, G. & Church, G. M. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 325, 1128–1131 (2009).
D'Costa, V. M., McGrann, K. M., Hughes, D. W. & Wright, G. D. Sampling the antibiotic resistome. Science 311, 374–377 (2006).
Ghosh, T. S., Gupta, S. S., Nair, G. B. & Mande, S. S. In silico analysis of antibiotic resistance genes in the gut microflora of individuals from diverse geographies and age-groups. PLoS ONE 8, e83823 (2013).
Hu, Y. et al. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nature Commun. 4, 2151 (2013).
Durso, L. M., Miller, D. N. & Wienhold, B. J. Distribution and quantification of antibiotic resistant genes and bacteria across agricultural and non-agricultural metagenomes. PLoS ONE 7, e48325 (2012).
D'Costa, V. M. et al. Antibiotic resistance is ancient. Nature 477, 457–461 (2011).
Finley, R. L. et al. The scourge of antibiotic resistance: the important role of the environment. Clin. Infect. Dis. 57, 704–710 (2013).
Ferrandiz, M. J., Fenoll, A., Linares, J. & De La Campa, A. G. Horizontal transfer of parC and gyrA in fluoroquinolone-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44, 840–847 (2000).
Baquero, F., Alvarez-Ortega, C. & Martinez, J. L. Ecology and evolution of antibiotic resistance. Environ. Microbiol. Rep. 1, 469–476 (2009).
Martinez, J. L. & Baquero, F. Mutation frequencies and antibiotic resistance. Antimicrob. Agents Chemother. 44, 1771–1777 (2000).
Laxminarayan, R. et al. Antibiotic resistance — the need for global solutions. Lancet Infect. Dis. 13, 1057–1098 (2013).
Bush, K. et al. Tackling antibiotic resistance. Nature Rev. Microbiol. 9, 894–896 (2011).
Baquero, F. Low-level antibacterial resistance: a gateway to clinical resistance. Drug Resist. Updat. 4, 93–105 (2001).
Baquero, F. European standards for antibiotic susceptibility testing: towards a theoretical consensus. Eur. J. Clin. Microbiol. Infect. Dis. 9, 492–495 (1990).
Kronvall, G. Normalized resistance interpretation as a tool for establishing epidemiological MIC susceptibility breakpoints. J. Clin. Microbiol. 48, 4445–4452 (2010).
Kahlmeter, G. et al. European harmonization of MIC breakpoints for antimicrobial susceptibility testing of bacteria. J. Antimicrob. Chemother. 52, 145–148 (2003).
Simjee, S., Silley, P., Werling, H. O. & Bywater, R. Potential confusion regarding the term 'resistance' in epidemiological surveys. J. Antimicrob. Chemother. 61, 228–229 (2008).
Morrissey, I. et al. Evaluation of epidemiological cut-off values indicates that biocide resistant subpopulations are uncommon in natural isolates of clinically-relevant microorganisms. PLoS ONE 9, e86669 (2014).
Garcia-Leon, G., Salgado, F., Oliveros, J. C., Sanchez, M. B. & Martinez, J. L. Interplay between intrinsic and acquired resistance to quinolones in Stenotrophomonas maltophilia. Environ. Microbiol. 16, 1282–1296 (2014).
Alvarez-Ortega, C., Wiegand, I., Olivares, J., Hancock, R. E. & Martinez, J. L. Genetic determinants involved in the susceptibility of Pseudomonas aeruginosa to ß-lactam antibiotics. Antimicrob. Agents Chemother. 54, 4159–4167 (2010).
Girgis, H. S., Hottes, A. K. & Tavazoie, S. Genetic architecture of intrinsic antibiotic susceptibility. PLoS ONE 4, e5629 (2009).
Fajardo, A. et al. The neglected intrinsic resistome of bacterial pathogens. PLoS ONE 3, e1619 (2008).
Fernandez, L. et al. Characterization of the polymyxin B resistome of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 57, 110–119 (2013).
Forsberg, K. J. et al. Bacterial phylogeny structures soil resistomes across habitats. Nature 509, 612–616 (2014).
Parsley, L. C. et al. Identification of diverse antimicrobial resistance determinants carried on bacterial, plasmid, or viral metagenomes from an activated sludge microbial assemblage. Appl. Environ. Microbiol. 76, 3753–3757 (2010).
Zankari, E. et al. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644 (2012).
Scaria, J., Chandramouli, U. & Verma, S. K. Antibiotic Resistance Genes Online (ARGO): a database on vancomycin and ß-lactam resistance genes. Bioinformation 1, 5–7 (2005).
Modi, S. R., Lee, H. H., Spina, C. S. & Collins, J. J. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature 499, 219–222 (2013).
Liu, B. & Pop, M. ARDB — Antibiotic Resistance Genes Database. Nucleic Acids Res. 37, D443–D447 (2009).
Forslund, K. et al. Country-specific antibiotic use practices impact the human gut resistome. Genome Res. 23, 1163–1169 (2013).
Davies, J. Inactivation of antibiotics and the dissemination of resistance genes. Science 264, 375–382 (1994).
Benveniste, R. & Davies, J. Aminoglycoside antibiotic-inactivating enzymes in actinomycetes similar to those present in clinical isolates of antibiotic-resistant bacteria. Proc. Natl Acad. Sci. USA 70, 2276–2280 (1973).
Martinez, J. L. et al. A global view of antibiotic resistance. FEMS Microbiol. Rev. 33, 44–65 (2009).
Martinez, J. L. et al. Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol. Rev. 33, 430–449 (2009).
Piddock, L. J. Multidrug-resistance efflux pumps — not just for resistance. Nature Rev. Microbiol. 4, 629–636 (2006).
Garcia-Leon, G. et al. A function of SmeDEF, the major quinolone resistance determinant of Stenotrophomonas maltophilia, is the colonization of the roots of the plants. Appl. Environ. Microbiol. http://dx.doi.org/10.1128/AEM.01058-14 (2014).
Thanassi, D. G., Cheng, L. W. & Nikaido, H. Active efflux of bile salts by Escherichia coli. J. Bacteriol. 179, 2512–2518 (1997).
Henderson, T. A., Young, K. D., Denome, S. A. & Elf, P. K. AmpC and AmpH, proteins related to the class C ß-lactamases, bind penicillin and contribute to the normal morphology of Escherichia coli. J. Bacteriol. 179, 6112–6121 (1997).
Ayala, F. J. Adaptation and novelty: teleological explanations in evolutionary biology. Hist. Philos. Life Sci. 21, 3–33 (1999).
Laskaris, P., Tolba, S., Calvo-Bado, L. & Wellington, L. Coevolution of antibiotic production and counter-resistance in soil bacteria. Environ. Microbiol. 12, 783–796 (2010).
Hiramatsu, K. et al. Genomic basis for methicillin resistance in Staphylococcus aureus. Infect. Chemother. 45, 117–136 (2013).
Martinez, J. L. Natural antibiotic resistance and contamination by antibiotic resistance determinants: the two ages in the evolution of resistance to antimicrobials. Front. Microbiol. 3, 1 (2012).
Martinez, J. L. The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proc. Biol. Sci. 276, 2521–2530 (2009).
Martinez, J. L., Baquero, F. & Andersson, D. I. Predicting antibiotic resistance. Nature Rev. Microbiol. 5, 958–965 (2007).
Hachler, H., Cohen, S. P. & Levy, S. B. marA, a regulated locus which controls expression of chromosomal multiple antibiotic resistance in Escherichia coli. J. Bacteriol. 173, 5532–5538 (1991).
Li, X. Z., Nikaido, H. & Poole, K. Role of MexA–MexB –OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39, 1948–1953 (1995).
Costa, Y., Galimand, M., Leclercq, R., Duval, J. & Courvalin, P. Characterization of the chromosomal aac(6′)-Ii gene specific for Enterococcus faecium. Antimicrob. Agents Chemother. 37, 1896–1903 (1993).
Fevre, C. et al. Six groups of the OXY ß-lactamase evolved over millions of years in Klebsiella oxytoca. Antimicrob. Agents Chemother. 49, 3453–3462 (2005).
Dantas, G. & Sommer, M. O. Context matters — the complex interplay between resistome genotypes and resistance phenotypes. Curr. Opin. Microbiol. 15, 577–582 (2012).
Zscheck, K. K. & Murray, B. E. Genes involved in the regulation of ß-lactamase production in enterococci and staphylococci. Antimicrob. Agents Chemother. 37, 1966–1970 (1993).
Enne, V. I., Delsol, A. A., Roe, J. M. & Bennett, P. M. Evidence of antibiotic resistance gene silencing in Escherichia coli. Antimicrob. Agents Chemother. 50, 3003–3010 (2006).
Yong, D. et al. Characterization of a new metallo-ß-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054 (2009).
Picao, R. C. et al. Plasmid-mediated quinolone resistance in Aeromonas allosaccharophila recovered from a Swiss lake. J. Antimicrob. Chemother. 62, 948–950 (2008).
Jones, B. V. & Marchesi, J. R. Transposon-aided capture (TRACA) of plasmids resident in the human gut mobile metagenome. Nature Methods 4, 55–61 (2007).
Marcone, G. L. et al. Novel mechanism of glycopeptide resistance in the A40926 producer Nonomuraea sp. ATCC 39727. Antimicrob. Agents Chemother. 54, 2465–2472 (2010).
Gu, B., Kelesidis, T., Tsiodras, S., Hindler, J. & Humphries, R. M. The emerging problem of linezolid-resistant Staphylococcus. J. Antimicrob. Chemother. 68, 4–11 (2013).
Spanogiannopoulos, P., Waglechner, N., Koteva, K. & Wright, G. D. A rifamycin inactivating phosphotransferase family shared by environmental and pathogenic bacteria. Proc. Natl Acad. Sci. USA 111, 7102–7107 (2014).
Andersson, D. I. & Hughes, D. Persistence of antibiotic resistance in bacterial populations. FEMS Microbiol. Rev. 35, 901–911 (2011).
Andersson, D. I. & Hughes, D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nature Rev. Microbiol. 8, 260–271 (2010).
Brown, M. G. & Balkwill, D. L. Antibiotic resistance in bacteria isolated from the deep terrestrial subsurface. Microb. Ecol. 57, 484–493 (2009).
Warinner, C. et al. Pathogens and host immunity in the ancient human oral cavity. Nature Genet. 46, 336–344 (2014).
Davies, J. & Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74, 417–433 (2010).
Martinez, J. L. Bottlenecks in the transferability of antibiotic resistance from natural ecosystems to human bacterial pathogens. Front. Microbiol. 2, 265 (2011).
Aminov, R. I. & Mackie, R. I. Evolution and ecology of antibiotic resistance genes. FEMS Microbiol. Lett. 271, 147–161 (2007).
Skippington, E. & Ragan, M. A. Lateral genetic transfer and the construction of genetic exchange communities. FEMS Microbiol. Rev. 35, 707–735 (2011).
Cohen, O., Gophna, U. & Pupko, T. The complexity hypothesis revisited: connectivity rather than function constitutes a barrier to horizontal gene transfer. Mol. Biol. Evol. 28, 1481–1489 (2011).
Baquero, F., Tedim, A. P. & Coque, T. M. Antibiotic resistance shaping multi-level population biology of bacteria. Front. Microbiol. 4, 15 (2013).
Levin, B. R. & Bull, J. J. Short-sighted evolution and the virulence of pathogenic microorganisms. Trends Microbiol. 2, 76–81 (1994).
Sibold, C. et al. Mosaic pbpX genes of major clones of penicillin-resistant Streptococcus pneumoniae have evolved from pbpX genes of a penicillin-sensitive Streptococcus oralis. Mol. Microbiol. 12, 1013–1023 (1994).
Martinez, J. L., Baquero, F. & Andersson, D. I. Beyond serial passages: new methods for predicting the emergence of resistance to novel antibiotics. Curr. Opin. Pharmacol. 11, 439–445 (2011).
Olivares, J. et al. Overproduction of the multidrug efflux pump MexEF–OprN does not impair Pseudomonas aeruginosa fitness in competition tests, but produces specific changes in bacterial regulatory networks. Environ. Microbiol. 14, 1968–1981 (2012).
Sanchez, M. B. & Martinez, J. L. Differential epigenetic compatibility of qnr antibiotic resistance determinants with the chromosome of Escherichia coli. PLoS ONE 7, e35149 (2012).
Levin, B. R., Perrot, V. & Walker, N. Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics 154, 985–997 (2000).
Steinkraus, G., White, R. & Friedrich, L. Vancomycin MIC creep in non-vancomycin-intermediate Staphylococcus aureus (VISA), vancomycin-susceptible clinical methicillin-resistant S. aureus (MRSA) blood isolates from 2001–2005. J. Antimicrob. Chemother. 60, 788–794 (2007).
Gibson, M. K., Forsberg, K. J. & Dantas, G. Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J. http://dx.doi.org/10.1038/ismej.2014.106 (2014).
Acknowledgements
Research in the author's laboratories is funded by the European Commission (EvoTAR-282004 for JLM, TMC and FB), the Ministry of Economy and Competitiveness (BIO2011-25255 for JLM, PI12-01581 for TMC, PI10-02588 for FB, and NEXTMICRO for FB and TMC), the Regional Government of Madrid in Spain (PROMPT-S2010/BMD2414 for JLM and FB), and by the Spanish Network for Research on Infectious Diseases (REIPI RD12/0015 for JLM) and the Spanish Network for the Study of Plasmids and Extrachromosomal Elements (REDEEX, BFU2011-14145-E for TMC).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information S1 (table)
Antibiotic resistance gene databases (PDF 159 kb)
PowerPoint slides
Rights and permissions
About this article
Cite this article
Martínez, J., Coque, T. & Baquero, F. What is a resistance gene? Ranking risk in resistomes. Nat Rev Microbiol 13, 116–123 (2015). https://doi.org/10.1038/nrmicro3399
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro3399
This article is cited by
-
Plasmids, a molecular cornerstone of antimicrobial resistance in the One Health era
Nature Reviews Microbiology (2024)
-
Impacts of irrigation with treated livestock wastewater on the accumulation characteristic of ARGs in the farmland soil: a case study in Hohhot, China
Environmental Geochemistry and Health (2024)
-
Longitudinal metagenomic study reveals the dynamics of fecal antibiotic resistome in pigs throughout the lifetime
Animal Microbiome (2023)
-
Development of the oral resistome during the first decade of life
Nature Communications (2023)
-
Evidence for wastewaters as environments where mobile antibiotic resistance genes emerge
Communications Biology (2023)