Key Points
-
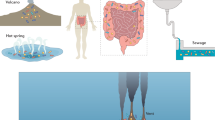
Microoxic environments are prevalent in nature, including in aquatic, terrestrial and host-associated environments.
-
Although a subset of bacteria known as microaerophiles grow optimally under microoxic conditions, many bacteria that have traditionally been characterized as aerobes or anaerobes also occupy these environments. All microorganisms that are capable of respiring low levels of O2 are defined as microaerobes.
-
Cytochrome oxidases with a high affinity for O2 are common in phylogenetically diverse bacterial genomes (∼70% of species surveyed) and provide microaerobes with access to scarce supplies of O2.
-
Analysis of shotgun metagenomes reveals a widespread occurrence of genes encoding high-affinity oxidases, providing intriguing evidence that supports the importance of microaerobic metabolism.
-
Understanding the physiology and ecology of microaerobes in low-O2 environments is integral to advancing our understanding of host-associated microbiomes, for both colonization and pathogen invasion.
-
Advances in both O2-sensing technologies and microoxic cultivation will provide opportunities for the exploration of the local and global impacts of microaerobes.
Abstract
Competition for molecular oxygen (O2) among respiratory microorganisms is intense because O2 is a potent electron acceptor. This competition leads to the formation of microoxic environments wherever microorganisms congregate in aquatic, terrestrial and host-associated communities. Bacteria can harvest O2 present at low, even nanomolar, concentrations using high-affinity terminal oxidases. Here, we report the results of surveys searching for high-affinity terminal oxidase genes in sequenced bacterial genomes and shotgun metagenomes. The results indicate that bacteria with the potential to respire under microoxic conditions are phylogenetically diverse and intriguingly widespread in nature. We explore the implications of these findings by highlighting the importance of microaerobic metabolism in host-associated bacteria related to health and disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Brochier-Armanet, C., Talla, E. & Gribaldo, S. The multiple evolutionary histories of dioxygen reductases: implications for the origin and evolution of aerobic respiration. Mol. Biol. Evol. 26, 285–297 (2009). This article includes genomic surveys of terminal oxidase genes and presents a model for the evolutionary origin of aerobic respiration in bacteria and archaea.
Stolper, D. A., Revsbech, N. P. & Canfield, D. E. Aerobic growth at nanomolar oxygen concentrations. Proc. Natl Acad. Sci. USA 107, 18755–18760 (2010). This study demonstrates aerobic growth of an E. coli strain at nanomolar concentrations of O 2 — concentrations that are orders of magnitude lower than those previously reported.
Marteyn, B. et al. Modulation of Shigella virulence in response to available oxygen in vivo. Nature 465, 355–358 (2010). This investigation of the effects of O 2 on virulence in Shigella includes the visualization of an oxygenated zone adjacent to the intestinal mucosa.
Lieberman, T. D. et al. Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nature Genet. 43, 1275–1280 (2011).
Abreu, I. A., Xavier, A. V., LeGall, J., Cabelli, D. E. & Teixeira, M. Superoxide scavenging by neelaredoxin: dismutation and reduction activities in anaerobes. J. Biol. Inorg. Chem. 7, 668–674 (2002).
Madigan, M. T., Martinko, J. M., Stahl, D. A. & Clark, D. P. Brock Biology of Microorganisms 13th edn 117–149 (Benjamin Cummings, 2010).
Baughn, A. D. & Malamy, M. H. The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature 427, 441–444 (2004).
Tiedje, J., Sexstone, A., Parkin, T., Revsbech, N. & Shelton, D. Anaerobic processes in soil. Plant Soils 76, 197–212 (1984).
Ploug, H., Iversen, M. H. & Fischer, G. Ballast, sinking velocity, and apparent diffusivity within marine snow and zooplankton fecal pellets: implications for substrate turnover by attached bacteria. Limnol. Oceanogr. 53, 1878–1886 (2008).
Alldredge, A. L. & Cohen, Y. Can microscale chemical patches persist in the sea? Microelectrode study of marine snow, fecal pellets. Science 235, 689–691 (1987).
Ploug, H. Small-scale oxygen fluxes and remineralization in sinking aggregates. Limnol. Oceanogr. 46, 1624–1631 (2001).
Stewart, P. S. & Franklin, M. J. Physiological heterogeneity in biofilms. Nature Rev. Microbiol. 6, 199–210 (2008).
Fenchel, T. & Finlay, B. Oxygen and the spatial structure of microbial communities. Biol. Rev. Camb. Philos. Soc. 83, 553–569 (2008).
Kühl, M., Rickelt, L. F. & Thar, R. Combined imaging of bacteria and oxygen in biofilms. Appl. Environ. Microbiol. 73, 6289–6295 (2007).
Wright, J. J., Konwar, K. M. & Hallam, S. J. Microbial ecology of expanding oxygen minimum zones. Nature Rev. Microbiol. 10, 381–394 (2012).
Soupène, E., Foussard, M., Boistard, P., Truchet, G. & Batut, J. Oxygen as a key developmental regulator of Rhizobium meliloti N2-fixation gene expression within the alfalfa root nodule. Proc. Natl Acad. Sci. USA 92, 3759–3763 (1995).
Kuzma, M. M., Hunt, S. & Layzell, D. B. Role of oxygen in the limitation and inhibition of nitrogenase activity and respiration rate in individual soybean nodules. Plant Physiol. 101, 161–169 (1993).
Charrier, M. & Brune, A. The gut microenvironment of helicid snails (Gastropoda: Pulmonata): in-situ profiles of pH, oxygen, and hydrogen determined by microsensors. Can. J. Zool. 81, 928–935 (2003).
Brune, A., Emerson, D. & Breznak, J. A. The termite gut microflora as an oxygen sink: microelectrode determination of oxygen and pH gradients in guts of lower and higher termites. Appl. Environ. Microbiol. 61, 2681–2687 (1995).
Van den Abbeele, P., Van de Wiele, T., Verstraete, W. & Possemiers, S. The host selects mucosal and luminal associations of coevolved gut microorganisms: a novel concept. FEMS Microbiol. Rev. 35, 681–704 (2011).
Mirza, B. S. & Rodrigues, J. L. Development of a direct isolation procedure for free-living diazotrophs under controlled hypoxic conditions. Appl. Environ. Microbiol. 16, 5542–5549 (2012).
Young, V. B. et al. Multiphasic analysis of the temporal development of the distal gut microbiota in patients following ileal pouch anal anastomosis. Microbiome (in the press).
Brioukhanov, A. & Netrusov, A. Aerotolerance of strictly anaerobic microorganisms and factors of defense against oxidative stress: a review. Appl. Biochem. Microbiol. 43, 567–582 (2007).
Hemp, J. et al. Evolutionary migration of a post-translationally modified active-site residue in the proton-pumping heme-copper oxygen reductases. Biochemistry 45, 15405–15410 (2006).
Calhoun, M. W., Thomas, J. W. & Gennis, R. B. The cytochrome oxidase superfamily of redox-driven proton pumps. Trends Biochem. Sci. 19, 325–330 (1994).
Pitcher, R. S. & Watmough, N. J. The bacterial cytochrome cbb 3 oxidases. Biochim. Biophys. Acta 1655, 388–399 (2004).
Brändén, G., Gennis, R. B. & Brzezinski, P. Transmembrane proton translocation by cytochrome c oxidase. Biochim. Biophys. Acta 1757, 1052–1063 (2006).
Puustinen, A., Finel, M., Haltia, T., Gennis, R. B. & Wikström, M. Properties of the two terminal oxidases of Escherichia coli. Biochemistry 30, 3936–3942 (1991).
Pereira, M. M., Santana, M. & Teixeira, M. A novel scenario for the evolution of haem–copper oxygen reductases. Biochim. Biophys. Acta 1505, 185–208 (2001).
Han, H. et al. Adaptation of aerobic respiration to low O2 environments. Proc. Natl Acad. Sci. USA 108, 14109–14114 (2011). This report discusses the implications of the adaptation of aerobic respiration to low-O 2 environments.
Pitcher, R. S., Brittain, T. & Watmough, N. J. Cytochrome cbb3 oxidase and bacterial microaerobic metabolism. Biochem. Soc. Trans. 30, 653–658 (2002).
Keightley, J. A. et al. Molecular genetic and protein chemical characterization of the cytochrome ba 3 from Thermus thermophilus HB8. J. Biol. Chem. 270, 20345–20358 (1995).
Rice, C. W. & Hempfling, W. P. Oxygen-limited continuous culture and respiratory energy conservation in Escherichia coli. J. Bacteriol. 134, 115–124 (1978).
D'mello, R., Hill, S. & Poole, R. K. The cytochrome bd quinol oxidase in Escherichia coli has an extremely high oxygen affinity and two oxygen-binding haems: implications for regulation of activity in vivo by oxygen inhibition. Microbiology 142, 755–763 (1996).
Jackson, R. J. et al. Oxygen reactivity of both respiratory oxidases in Campylobacter jejuni: the cydAB genes encode a cyanide-resistant, low-affinity oxidase that is not of the cytochrome bd type. J. Bacteriol. 189, 1604–1615 (2007).
Cunningham, L., Pitt, M. & Williams, H. D. The cioAB genes from Pseudomonas aeruginosa code for a novel cyanide-insensitive terminal oxidase related to the cytochrome bd quinol oxidases. Mol. Microbiol. 24, 579–591 (1997).
Arai, H. Regulation and function of versatile aerobic and anaerobic respiratory metabolism in Pseudomonas aeruginosa. Front. Microbiol. 2, 103 (2011).
Altschul, S., Gish, W., Miller, W., Myers, E. & Lipman, D. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Ducluzeau, A. L., Ouchane, S. & Nitschke, W. The cbb 3 oxidases are an ancient innovation of the domain Bacteria. Mol. Biol. Evol. 25, 1158–1166 (2008).
Pace, N. R. A molecular view of microbial diversity and the biosphere. Science 276, 734–740 (1997).
Meyer, F. et al. The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9, 386 (2008).
Giovannoni, S. J. et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science 309, 1242–1245 (2005).
Borisov, V. B. et al. Redox control of fast ligand dissociation from Escherichia coli cytochrome bd. Biochem. Biophys. Res. Commun. 355, 97–102 (2007).
Reinders, C. A. et al. Rectal nitric oxide and fecal calprotectin in inflammatory bowel disease. Scand. J. Gastroenterol. 42, 1151–1157 (2007).
Borisov, V. B. et al. Aerobic respiratory chain of Escherichia coli is not allowed to work in fully uncoupled mode. Proc. Natl Acad. Sci. USA 108, 17320–17324 (2011).
Wertz, J. T. & Breznak, J. A. Physiological ecology of Stenoxybacter acetivorans, an obligate microaerophile in termite guts. Appl. Environ. Microbiol. 73, 6829–6841 (2007).
Preisig, O., Zufferey, R., Thöny-Meyer, L., Appleby, C. A. & Hennecke, H. A high-affinity cbb 3-type cytochrome oxidase terminates the symbiosis-specific respiratory chain of Bradyrhizobium japonicum. J. Bacteriol. 178, 1532–1538 (1996).
Wiles, S., Pickard, K. M., Peng, K., MacDonald, T. T. & Frankel, G. In vivo bioluminescence imaging of the murine pathogen Citrobacter rodentium. Infect. Immun. 74, 5391–5396 (2006).
Goldman, B. S., Gabbert, K. K. & Kranz, R. G. The temperature-sensitive growth and survival phenotypes of Escherichia coli cydDC and cydAB strains are due to deficiencies in cytochrome bd and are corrected by exogenous catalase and reducing agents. J. Bacteriol. 178, 6348–6351 (1996).
Jones, S. A. et al. Respiration of Escherichia coli in the mouse intestine. Infect. Immun. 75, 4891–4899 (2007).
Jones, S. A. et al. Anaerobic respiration of Escherichia coli in the mouse intestine. Infect. Immun. 79, 4218–4226 (2011).
Weingarten, R. A., Grimes, J. L. & Olson, J. W. Role of Campylobacter jejuni respiratory oxidases and reductases in host colonization. Appl. Environ. Microbiol. 74, 1367–1375 (2008).
Way, S. S., Sallustio, S., Magliozzo, R. S. & Goldberg, M. B. Impact of either elevated or decreased levels of cytochrome bd expression on Shigella flexneri virulence. J. Bacteriol. 181, 1229–1237 (1999).
Alvarez-Ortega, C. & Harwood, C. S. Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol. Microbiol. 65, 153–165 (2007).
Shi, L. et al. Changes in energy metabolism of Mycobacterium tuberculosis in mouse lung and under in vitro conditions affecting aerobic respiration. Proc. Natl Acad. Sci. USA 102, 15629–15634 (2005).
Battino, R., Rettich, T. R. & Tominaga, T. The solubility of oxygen and ozone in liquids. J. Phys. Chem. Ref. Data 12, 163–178 (1983).
Revsbech, N. P., Thamdrup, B., Dalsgaard, T. & Canfield, D. E. Construction of STOX oxygen sensors and their application for determination of O2 concentrations in oxygen minimum zones. Methods Enzymol. 486, 325–341 (2011).
Klimant, I., Kuhl, M., Glud, R. N. & Holst, G. Optical measurement of oxygen and temperature in micro scale: strategies and biological applications. Sens. Actuators B Chem. 38, 29–37 (1997).
Grate, J. W., Kelly, R. T., Suter, J. & Anheier, N. C. Silicon-on-glass pore network micromodels with oxygen-sensing fluorophore films for chemical imaging and defined spatial structure. Lab Chip 12, 4796–4801 (2012).
Røy, H. et al. Aerobic microbial respiration in 86-million-year-old deep-sea red clay. Science 336, 922–925 (2012).
Acknowledgements
The authors thank J. Breznak for seminal conversations about the ecological importance of microaerobes; K. D. Noel and C. Waldron for critical reading of this manuscript; T. Teal and B. Klahn for their expert assistance with the genomic and metagenomic analyses; and members of the Schmidt laboratory for many helpful discussions concerning the potential consequences of microaerobes and microoxic environments. This work was supported by grants from the US National Institutes of Health (R01 HG004906 and UH3 DK083993) and the US National Science Foundation (MCB 0731913 and DEB 1027253).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary informtation S1 (table)
Occurrence of high-affinity oxidase genes in bacterial genomes. (PDF 201 kb)
Supplementary informtation S2 (box)
Metagenome Analysis of Terminal Oxidase Genes (PDF 219 kb)
Supplementary informtation S3 (table)
MG-RAST accession numbers for the metagenomes in Figures 2 and 3. (PDF 201 kb)
Related links
Rights and permissions
About this article
Cite this article
Morris, R., Schmidt, T. Shallow breathing: bacterial life at low O2. Nat Rev Microbiol 11, 205–212 (2013). https://doi.org/10.1038/nrmicro2970
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro2970
This article is cited by
-
Genomic insight of sulfate reducing bacterial genus Desulfofaba reveals their metabolic versatility in biogeochemical cycling
BMC Genomics (2023)
-
Anaerobic thiosulfate oxidation by the Roseobacter group is prevalent in marine biofilms
Nature Communications (2023)
-
An in vitro platform for study of the human gut microbiome under an oxygen gradient
Biomedical Microdevices (2023)
-
New globally distributed bacterial phyla within the FCB superphylum
Nature Communications (2022)
-
Identification of nosZ-expressing microorganisms consuming trace N2O in microaerobic chemostat consortia dominated by an uncultured Burkholderiales
The ISME Journal (2022)