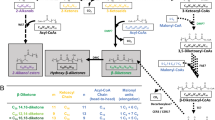
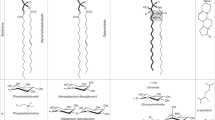
Key Points
-
Hopanes were discovered by petroleum geologists as ubiquitous molecular fossils in ancient sedimentary rocks. Later, bacterial hopanoids were identified as their progenitors.
-
Today, phylogenetically diverse bacteria make hopanoids using machinery encoded by a conserved set of genes. The rhizosphere appears to be a niche that is common to many hopanoid-producing bacteria, and some hopanoid producers are known plant symbionts.
-
Bacteria make structurally distinct types of hopanoids, including ones that can covalently bind lipid A. Different hopanoid classes exhibit different properties and likely have specific biological functions.
-
Hopanoids share similar biophysical properties with sterols, such as tuning membrane rigidity and permeability. Though some evidence suggests that hopanoids help order membranes, how such ordering impacts cells and whether hopanoids interact with particular membrane proteins remain to be determined.
-
In vitro, hopanoids contribute to bacterial stress resistance, which may help explain their ability to facilitate beneficial plant–bacteria interactions. However, given that hopanoids can also serve as carriers for plant hormones and that plants themselves make hopanoid-like compounds, it is likely that other mechanisms are additionally at play.
Abstract
Lipid research represents a frontier for microbiology, as showcased by hopanoid lipids. Hopanoids, which resemble sterols and are found in the membranes of diverse bacteria, have left an extensive molecular fossil record. They were first discovered by petroleum geologists. Today, hopanoid-producing bacteria remain abundant in various ecosystems, such as the rhizosphere. Recently, great progress has been made in our understanding of hopanoid biosynthesis, facilitated in part by technical advances in lipid identification and quantification. A variety of genetically tractable, hopanoid-producing bacteria have been cultured, and tools to manipulate hopanoid biosynthesis and detect hopanoids are improving. However, we still have much to learn regarding how hopanoid production is regulated, how hopanoids act biophysically and biochemically, and how their production affects bacterial interactions with other organisms, such as plants. The study of hopanoids thus offers rich opportunities for discovery.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Fahy, E., Sud, M., Cotter, D. & Subramaniam, S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 35, W606–W612 (2007).
Rohmer, M., Bouvier, P. & Ourisson, G. Molecular evolution of biomembranes: structural equivalents and phylogenetic precursors of sterols. Proc. Natl Acad. Sci. USA 76, 847–851 (1979).
Burlingame, A. L., Haug, P. & Belsky, T. Calvin, M. Occurrence of biogenic steranes and pentacyclic triterpanes in an Eocene shale (52 millions years) and in an early Pre-Cambrian shale (2.7 billion years): a preliminary report. Proc. Natl Acad. Sci. USA 54, 1406–1412 (1965).
Brocks, J. J. & Banfield, J. Biomarker evidence for green and purple sulphur bacteria in a stratified Paleoproterozoic sea. Nature 437, 866–870 (2005).
Ourisson, G. & Albrecht, P. Geohopanoids: the most abundant natural products on Earth? Acc. Chem. Res. 25, 398–402 (1992).
Bird, C. W., Lynch, J. M., Pirt, S. J. & Reid, W. W. The identification of hop-22(29)-ene in prokaryotic organisms. Tetrahedron Lett. 12, 3189–3190 (1971).
De Rosa, M., Gambacorta, A., Minale, L. & Bu'Lock, J. D. Bacterial triterpenes. J. Chem. Soc. D Chem. Commun., 619–620 (1971). References 6 and 7 were the first papers to discover hopanoids in bacteria.
Newman, D. K., Neubauer, C., Ricci, J. N., Wu, C.-H. & Pearson, A. Cellular and molecular biological approaches to interpreting ancient biomarkers. Annu. Rev. Earth Planet. Sci. 44, 493–522 (2016).
Pearson, A. in Treatise on Geochemistry 2nd edn (eds Falkowski, P. G. & Freeman, K. H.) 291–336. (Elsevier, London, 2014).
Vranová, E., Coman, D. & Gruissem, W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 64, 665–700 (2013).
Abe, I. Enzymatic synthesis of cyclic triterpenes. Nat. Prod. Rep. 24, 1311–1321 (2007).
Christianson, D. W. Structural biology and chemistry of the terpenoid cyclases. Chem. Rev. 106, 3412–3442 (2006).
Barton, D. H. et al. Biosynthesis of 3beta-hydroxy-triterpenoids and -steroids from (3S)-2,3-epoxy-2,3-dihydrosqualene. J. Chem. Soc. Perkin Trans. 1 12, 1134–1138 (1975).
Wei, J. H., Yin, X. & Welander, P. V. Sterol synthesis in diverse Bacteria. Front. Microbiol. 7, 259–219 (2016).
Rohmer, M., Anding, C. & Ourisson, G. Non-specific biosynthesis of hopane triterpenes by a cell-free system from Acetobacter pasteurianum. Eur. J. Biochem. 112, 541–547 (1980).
Poralla, K. The possible role of a repetitive amino acid motif in evolution of triterpenoid cyclases. Bioorg. Med. Chem. Lett. 4, 285–290 (1994).
Ourisson, G. & Nakatani, Y. The terpenoid theory of the origin of cellular life: the evolution of terpenoids to cholesterol. Chem. Biol. 1, 11–23 (1994).
Syrén, P.-O., Henche, S., Eichler, A., Nestl, B. M. & Hauer, B. Squalene-hopene cyclases — evolution, dynamics and catalytic scope. Curr. Opin. Struct. Biol. 41, 73–82 (2016).
Ourisson, G., Albrecht, P. & Rohmer, M. Predictive microbial biochemistry — from molecular fossils to procaryotic membranes. Trends Biochem. Sci. 7, 236–239 (1982).
Fischer, W. W. & Pearson, A. Hypotheses for the origin and early evolution of triterpenoid cyclases. Geobiology 5, 19–34 (2007).
Banta, A. B., Wei, J. H. & Welander, P. V. A distinct pathway for tetrahymanol synthesis in bacteria. Proc. Natl Acad. Sci. USA 112, 13478–13483 (2015).
Fischer, W. W., Summons, R. E. & Pearson, A. Targeted genomic detection of biosynthetic pathways: anaerobic production of hopanoid biomarkers by a common sedimentary microbe. Geobiology 3, 33–40 (2005).
Ourisson, G., Albrecht, P. & Rohmer, M. The hopanoids: palaeochemistry and biochemistry of a group of natural products. Pure Appl. Chem. 51, 709–729 (1979). This paper is a concise compendium of key early references that describe the discovery of fossil hopanes in sedimentary rocks and their origin as bacterial hopanoids.
Racolta, S., Juhl, P. B., Sirim, D. & Pleiss, J. The triterpene cyclase protein family: a systematic analysis. Proteins 80, 2009–2019 (2012).
Devon, T. K. & Scott, A. I. in Handbook of Naturally Occurring Compounds Vol. 2 281–389 (Academic Press, New York, 1972).
Inayama, S., Hori, H. & Pang, G. M. Isolation of a hopane-type triterpenoid, zeorin, from a higher plant, Tripterygium regelii. Chem. Pharm. Bull. 37, 2836–2837 (1989). This work provides a short review of hopanoid-like triterpenoids found in plants and lichens.
Shinozaki, J., Shibuya, M., Masuda, K. & Ebizuka, Y. Squalene cyclase and oxidosqualene cyclase from a fern. FEBS Lett. 582, 310–318 (2008).
Frickey, T. & Kannenberg, E. Phylogenetic analysis of the triterpene cyclase protein family in prokaryotes and eukaryotes suggests bidirectional lateral gene transfer. Environ. Microbiol. 11, 1224–1241 (2009).
Lopes, D., Villela, C. T., Mac Kaplan & Carauta, J. Moretenolactone, a β-lactone hopanoid from Ficus insipida. Phytochem 34, 279–280 (1993).
Perzl, M. et al. Cloning of conserved genes from Zymomonas mobilis and Bradyrhizobium japonicum that function in the biosynthesis of hopanoid lipids. Biochim. Biophys. Acta 1393, 108–118 (1998).
Bradley, A. S., Pearson, A., Sáenz, J. P. & Marx, C. J. Adenosylhopane: The first intermediate in hopanoid side chain biosynthesis. Org. Geochem. 41, 1075–1081 (2010).
Flesch, G. & Rohmer, M. Prokaryotic hopanoids: the biosynthesis of the bacteriohopane skeleton. Formation of isoprenic units from two distinct acetate pools and a novel type of carbon/carbon linkage between a triterpene and D-ribose. Eur. J. Biochem. 175, 405–411 (1988).
Rohmer, M., Sutter, B. & Sahm, H. Bacterial sterol surrogates. Biosynthesis of the side-chain of bacteriohopanetetrol and of a carbocyclic pseudopentose from 13C-labelled glucose in Zymomonas mobilis. J. Chem. Soc. Chem. Commun. 19, 1471–1472 (1989).
Welander, P. V. et al. Identification and characterization of Rhodopseudomonas palustris TIE-1 hopanoid biosynthesis mutants. Geobiology 10, 163–177 (2012). This work shows that hpnH is essential for formation of all extended hopanoids.
Liu, W. et al. Ribosylhopane, a novel bacterial hopanoid, as precursor of C35 bacteriohopanepolyols in Streptomyces coelicolor A3(2). ChemBioChem 15, 2156–2161 (2014).
Schmerk, C. L. et al. Elucidation of the Burkholderia cenocepacia hopanoid biosynthesis pathway uncovers functions for conserved proteins in hopanoid-producing bacteria. Environ. Microbiol. 17, 735–750 (2014). This work identifies hopanoid biosynthesis-associated glycosyltransferase protein HpnI, hopanoid biosynthesis-associated radical SAM protein HpnJ and hopanoid biosynthesis-associated protein HpnK as enzymes that modify extended hopanoid tails.
Welander, P. V., Coleman, M. L., Sessions, A. L., Summons, R. E. & Newman, D. K. Identification of a methylase required for 2-methylhopanoid production and implications for the interpretation of sedimentary hopanes. Proc. Natl Acad. Sci. USA 19, 8537–8542 (2010).
Welander, P. V. & Summons, R. E. Discovery, taxonomic distribution, and phenotypic characterization of a gene required for 3-methylhopanoid production. Proc. Natl Acad. Sci. USA 109, 12905–12910 (2012).
Ricci, J. N., Michel, A. J. & Newman, D. K. Phylogenetic analysis of HpnP reveals the origin of 2-methylhopanoid production in Alphaproteobacteria. Geobiology 13, 267–277 (2015).
Rohmer, M., Bouvier, P. & Ourisson, G. Non-specific lanosterol and hopanoid biosynthesis by a cell-free system from the bacterium Methylococcus capsulatus. Eur. J. Biochem. 112, 557–560 (1980).
Herrmann, D. A non-extractable triterpenoid of the hopane series in Acetobacter xylinum. FEMS Microbiol. Lett. 135, 323–326 (1996).
Cvejic, J. et al. Bacterial triterpenoids of the hopane series as biomarkers for the chemotaxonomy of Burkholderia. Pseudomonas and Ralstonia spp. FEMS Microbiol. Lett. 183, 295–299 (2000).
Cvejic, J. et al. Bacterial triterpenoids of the hopane series from the methanotrophic bacteria Methylocaldum spp.: phylogenetic implications and first evidence for an unsaturated aminobacteriohopanepolyol. FEMS Microbiol. Lett. 182, 361–365 (2000).
Rohmer, M. & Ourisson, G. Unsaturated bacteriohopanepolyols from Acetobacter aceti ssp. xylinum. J. Chem. Res. (S), 356–357; (M), 3037–3059 (1976).
Bligh, E. & Dyer, W. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 (1959).
Sessions, A. L. et al. Identification and quantification of polyfunctionalized hopanoids by high temperature gas chromatography-mass spectrometry. Org. Geochem. 56, 120–130 (2013). This work establishes an optimized protocol for identification of structurally diverse hopanoids by gas chromatography.
Talbot, H. et al. Analysis of intact bacteriohopanepolyols from methanotrophic bacteria by reversed-phase high performance liquid chromatography-atmospheric pressure chemical ionization mass spectrometry. J. Chromatogr. A 921, 175–185 (2001). This work establishes an optimized protocol for identification of various extended hopanoids by high-performance liquid chromatography.
Wu, C.-H. et al. Quantitative hopanoid analysis enables robust pattern detection and comparison between laboratories. Geobiology 13, 391–407 (2015).
Tahara, Y., Yamashita, T., Kondo, M. & Yamada, Y. Isolation of Zymomonas mobilis mutant deficient in tetrahydroxybacteriohopane biosynthesis. Agr. Biol. Chem. 52, 3189–3190 (1988).
Jürgens, U. J., Simonin, P. & Rohmer, M. Localization and distribution of hopanoids in membrane systems of the cyanobacterium Synechocystis PCC 6714. FEMS Microbiol. Lett. 1097, 90723–90722 (1992).
Kleemann, G., Alskog, G., Berry, A. M. & Huss-Danell, K. Lipid composition and nitrogenase activity of symbiotic Frankia (Alnus incana) in response to different oxygen concentrations. Protoplasma 183, 107–115 (1994).
Doughty, D. M., Hunter, R. C., Summons, R. E. & Newman, D. K. 2-Methylhopanoids are maximally produced in akinetes of Nostoc punctiforme: geobiological implications. Geobiology 7, 524–532 (2009).
Wu, C.-H., Bialecka-Fornal, M. & Newman, D. K. Methylation at the C-2 position of hopanoids increases rigidity in native bacterial membranes. eLife 4, e05663 (2015). This work describes the specific effects of 2Me-hopanoids on the fluidity of membranes and compares the contribution of hopanoids to fluidity among inner, outer and native membranes.
Doughty, D. M. et al. The RND-family transporter, HpnN, is required for hopanoid localization to the outer membrane of Rhodopseudomonas palustris TIE-1. Proc. Natl Acad. Sci. USA 108, E1045–1051 (2011).
Kumar, N. et al. Crystal structures of the Burkholderia multivorans hopanoid transporter HpnN. Proc. Natl Acad. Sci. USA 114, 6557–6562 (2017).
Sandhu, P. & Akhter, Y. Evolution of structural fitness and multifunctional aspects of mycobacterial RND family transporters. Arch. Microbiol. http://doi.org/10.1007/s00203-017-1434-6 (2017).
Kannenberg, E., Poralla, K. & Blume, A. A hopanoid from the thermo-acidophilic Bacillus acidocaldarius condenses membranes. Naturwissenschaften 67, 458 (1980).
Poralla, K., Kannenberg, E. & Blume, A. A glycolipid containing hopane isolated from the acidophilic, thermophilic Bacillus acidocaldarius, has a cholesterol-like function in membranes. FEBS Lett. 113, 107–110 (1980). This work provides the first evidence that hopanoids can condense and increase the stability of lipid monolayers similarly to cholesterol.
Kannenberg, E., Blume, A. McElhaney, R. N. & Poralla, K. Monolayer and calorimetric studies of phosphatidylcholines containing branched-chain fatty acids and of their interactions with cholesterol and with a bacterial hopanoid in model membranes. Biochim. Biophys. Acta Biomembr. 733, 111–116 (1983).
Leonard, A. & Milon, A. Modulation of membrane hydrophobic thickness by cholesterol, cycloartenol and hopanoid. A solid state 2H-NMR Study. Bull. Magn. Reson. 15, 124–127 (1993).
Nagimo, A., Sato, Y. & Suzuki, Y. Electron spin resonance studies of phosphatidylcholine interacted with cholesterol and with a hopanoid in liposomal membrane. Chem. Pharm. Bull. 39, 3071–3074 (1991).
Chen, Z., Tanno, N. & Takenaka, S. Effects of bacteriohopane-32-ol on the stability of various kinds of liposomal membranes. Biol. Pharm. Bull. 18, 600–604 (1995).
Sato, Y., Chen, Z. & Suzuki, Y. Thermodynamic effects of hopanoids on synthetic and bacterial phospholipid membranes. Chem. Pharm. Bull. 43, 1241–1244 (1995).
Chen, Z., Sato, Y., Nakazawa, I. & Suzuki, Y. Interactions between bacteriohopane-32, 33, 34, 35-tetrol and liposomal membranes composed of dipalmitoylphosphatidylcholine. Biol. Pharm. Bull. 18, 477–480 (1995).
Kannenberg, E. & Poralla, K. The influence of hopanoids on growth of Mycoplasma mycoides. Arch. Microbiol. 133, 100 (1983).
Stroeve, P., Pettit, C. D., Vasquez, V., Kim, I. & Berry, A. M. Surface active behavior of hopanoid lipids: bacteriohopanetetrol and phenylacetate monoester bacteriohopanetetrol. Langmuir 14, 4261–4265 (1998).
Poger, D. & Mark, A. E. The relative effect of sterols and hopanoids on lipid bilayers: when comparable is not identical. J. Phys. Chem. B. 117, 16129–16140 (2013).
Neubauer, C. et al. Lipid remodeling in Rhodopseudomonas palustris TIE-1 upon loss of hopanoids and hopanoid methylation. Geobiology 13, 443–453 (2015).
Bradley, A. S. et al. Hopanoid-free Methylobacterium extorquens DM4 overproduces carotenoids and has widespread growth impairment. PLoS ONE 12, e0173323 (2017).
Gruszecki, W. I. & Strzalka, K. Carotenoids as modulators of lipid membrane physical properties. Biochim. Biophys. Acta 1740, 108–115 (2005).
Pike, L. J. The challenge of lipid rafts. J. Lipid Res. 50 (Suppl.), S323–S328 (2009).
Gumí- Audenis, B. et al. Structure and nanomechanics of model membranes by atomic force microscopy and spectroscopy: insights into the role of cholesterol and sphingolipids. Membranes 6, 58 (2016).
Brown, D. A. & London, E. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J. Biol. Chem. 275, 17221–17224 (2000).
Sezgin, E., Levental, I., Mayor, S. & Eggeling, C. The mystery of membrane organization: composition, regulation, and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 18, 361–374 (2017).
Farnoud, A. M., Toledo, A. M., Konopka, J. B., Del Poeta, M. & London, E. Raft-like membrane domains in pathogenic microorganisms. Curr. Top. Membr. 75, 233–268 (2015).
Bramkamp, M. & Lopez, D. Exploring the existence of lipid rafts in bacteria. Microbiol. Mol. Biol. Rev. 9, 81–100 (2015).
Strahl, H. & Errington, J. Bacterial membranes: structure, domains and function. Annu. Rev. Microbiol. 71, 519 (2017).
Sáenz, J. P., Sezgin, E., Schwille, P. & Simons, K. Functional convergence of hopanoids and sterols in membrane ordering. Proc. Natl Acad. Sci. USA 109, 14236–14240 (2012). This work demonstrates that hopanoids can confer membrane order via formation of liquid-ordered states in synthetic liposomes.
Sáenz, J. P. et al. Hopanoids as functional analogues of cholesterol in bacterial membranes. Proc. Natl Acad. Sci. USA, 112, 11971–11976 (2015). This work demonstrates that hopanoids facilitate ordering of bacterial outer membranes via interaction with saturated lipids such as lipid A as well as via enhancing multidrug efflux.
Sáenz, J. P. Hopanoid enrichment in a detergent resistant membrane fraction of Crocosphaera watsonii: implications for bacterial lipid raft formation. Org. Geochem. 41, 853–856 (2010).
Doughty, D. M., Dieterle, M., Sessions, A. L., Fischer, W. W. & Newman, D. K. Probing the subcellular localization of hopanoid lipids in bacteria using nanoSIMS. PLoS ONE 9, e84455 (2014).
Flesch, G. & Rohmer, M. Growth inhibition of hopanoid synthesizing bacteria by squalene cyclase inhibitors. Arch. Microbiol. 147, 100–104 (1987).
Horbach, S., Neuss, B. & Sahm, H. Effect of azasqualene on hopanoid biosynthesis and ethanol tolerance of Zymomonas mobilis. FEMS Microbiol. Lett. 79, 347–350 (1991).
Schmerk, C. L., Bernards, M. A. & Valvano, M. A. Hopanoid production is required for low-pH tolerance, antimicrobial resistance, and motility in Burkholderia cenocepacia. J. Bacteriol. 193, 6712–6723 (2011).
Seipke, R. F. & Loria, R. Hopanoids are not essential for growth of Streptomyces scabies 87–22. J. Bacteriol. 191, 5216–5223 (2009).
Kulkarni, G. & Busset, N. et al. Specific hopanoid classes differentially affect free-living and symbiotic states of Bradyrhizobium diazoefficiens. mBio 6, e01251-15 (2015). This work identifies extended hopanoids as the most important hopanoid structural class for stress resistance and symbiosis of Bradyrhizobium diazoefficiens.
Welander, P. V. et al. Hopanoids play a role in membrane integrity and pH homeostasis in Rhodopseudomonas palustris TIE-1. J. Bacteriol. 191, 6145–6156 (2009).
Poralla, K., Härtner, T. & Kannenberg, E. Effect of temperature and pH on the hopanoid content of Bacillus acidocaldarius. FEMS Microbiol. Lett. 23, 253–256 (1984).
Kulkarni, G., Wu, C.-H. & Newman, D. K. The general stress response factor EcfG regulates expression of the C-2 hopanoid methylase HpnP in Rhodopseudomonas palustris TIE-1. J. Bacteriol. 195, 2490–2498 (2013).
Ricci, J. N., Morton, R., Kulkarni, G., Summers, M. L. & Newman, D. K. Hopanoids play a role in stress tolerance and nutrient storage in the cyanobacterium Nostoc punctiforme. Geobiology 15, 173–183 (2016).
Garby, T. J. et al. Lack of methylated hopanoids renders the cyanobacterium Nostoc punctiforme sensitive to osmotic and pH Stress. Appl. Environ. Microbiol. 83, e00777-17 (2017).
Poralla, K., Muth, G. & Hartner, T. Hopanoids are formed during transition from substrate to aerial hyphae in Streptomyces coelicolor A3(2). FEMS Microbiol. Lett. 189, 93–95 (2000).
Bosak, T., Losick, R. M. & Pearson, A. A polycyclic terpenoid that alleviates oxidative stress. Proc. Natl Acad. Sci. USA 105, 6725–6729 (2008).
Caron, B., Mark, A. E. & Poger, D. Some like it hot: the effect of sterols and hopanoids on lipid ordering at high temperature. J. Phys. Chem. Lett. 5, 3953–3957 (2014).
Lopez, D. & Koch, G. Exploring functional membrane microdomains in bacteria: an overview. Curr. Opin. Microbiol. 36, 76–84 (2017).
Vilcheze, C., Llopiz, P. & Neunlist, S. Prokaryotic triterpenoids: new hopanoids from the nitrogen-fixing bacteria Azotobacter vinelandii. Beijerinckia indica and Beijerinckia mobilis. Microbiology 140, 2749–2753 (1994).
Rohmer, M., Bouvier-Nave, P. & Ourisson, G. Distribution of hopanoid triterpenes in prokaryotes. J. Gen. Microbiol. 130, 1137 (1984).
Kannenberg, E., Perzl, E. & Härtner, T. The occurrence of hopanoid lipids in Bradyrhizobium bacteria. FEMS Microbiol. Lett. 127, 255–261 (1995).
Rosa-Putra, S., Nalin, R., Domenach, A.-M. & Rohmer, M. Novel hopanoids from Frankia spp. and related soil bacteria: squalene cyclization and significance of geological biomarkers revisited. Eur. J. Biochem. 268, 4300 (2001).
Hakoyama, T. et al. Host plant genome overcomes the lack of a bacterial gene for symbiotic nitrogen fixation. Nature 462, 514–517 (2009).
Appleby, C. A. Leghemoglobin and rhizobium respiration. Annu. Rev. Plant Physiol. 35, 443–478 (1984).
Sabra, W., Zeng, A. P. & Lünsdorf, H. Effect of oxygen on formation and structure of Azotobacter vinelandii alginate and its role in protecting nitrogenase. Appl. Environ. Microbiol. 66, 4037–4044 (2000).
Parsons, R., Silvester, W., Harris, S., Gruijters, W. & Bullivant, S. Frankia vesicles provide inducible and absolute oxygen protection for nitrogenase. Plant Physiol. 83, 728–731 (1987).
Abeysekera, R. M., Newcomb, W., Silvester, W. B. & Torrey, J. G. A freeze-fracture electron microscopy study of Frankia in root nodules of Alnus incana grown at three oxygen tensions. Can. J. Microbiol. 36, 97–108 (1990).
Berry, A. M., Moreau, R. A. & Jones, A. D. Bacteriohopanetetrol: abundant lipid in Frankia cells and in nitrogen-fixing nodule tissue. Plant Physiol. 95, 111–115 (1991).
Fries, L. Growth regulating effects of phenylacetic acid and phydroxy-phenylacetic acid on Fucus spiralis L. (Phaecophyceae. Fucales) in axenic culture. Phycology 16, 451–455 (1977).
Hammad, Y. et al. A possible role for phenyl acetic acid (PAA) on Alnus glutinosa nodulation by Frankia. Plant Soil 254, 193 (2003). This work finds that the auxinomimetic plant hormone PAA can be released from PAA-conjugated hopanoid lipids to regulate nodulation in Frankia symbioses.
Ricci, J. N. et al. Diverse capacity for 2-methylhopanoid production correlates with a specific ecological niche. ISME J. 8, 675–684 (2013). This work identifies a important association between 2Me-hopanoids and plant-associated environments.
Kloepper, J. W., Ryu, C. M. & Zhang, S. Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology 94, 1259–1266 (2004).
Lemaire, B. et al. Biogeographical patterns of legume-nodulating Burkholderia spp.: from African Fynbos to continental scales. Appl. Environ. Microbiol. 82, 5099–5115 (2016).
Moulin, L., James, E. K., Klonowska, A., Daria, S. M. & Simon, M. F. in Biological Nitrogen Fixation (ed. de Bruijn, F. J.) 177–190 (John Wiley & Sons, Hoboken, NJ, USA, 2015).
Sy, A. et al. Methylotrophic Methylobacterium bacteria nodulate and fix nitrogen in symbiosis with legumes. J. Bacteriol. 183, 214–220 (2001).
Silipo, A. et al. Covalently linked hopanoid-lipid A improves outer-membrane resistance of a Bradyrhizobium symbiont of legumes. Nat. Commun. 5, 5106 (2014). This work describes a novel HoLA structure and provides the first evidence that hopanoids affect legume–rhizobia symbioses.
Delamuta, J. R. M. et al. Polyphasic evidence supporting the reclassification of Bradyrhizobium japonicum group Ia strains as Bradyrhizobium diazoefficiens sp. nov. Int. J. Syst. Evol. Microbiol. 63, 3342–3351 (2013).
Komaniecka, I. et al. Occurrence of an unusual hopanoid-containing lipid A among lipopolysaccharides from Bradyrhizobium species. J. Biol. Chem. 289, 35644–35655 (2014).
Busset, N. et al. The very long chain fatty acid (C26:25OH) linked to the lipid A Is important for the fitness of the photosynthetic Bradyrhizobium strain ORS278 and the establishment of a successful symbiosis with Aeschynomene legumes. Front. Microbiol. 8, 1821 (2017).
Shevchenko, A., Schwille, P. & Simons, K. Yeast lipids can phase-separate into micrometer-scale membrane domains. J. Biol. Chem. 285, 30224–30232 (2010).
Boyd, E. S., Hamilton, T. L., Wang, J., He, L. & Zhang, C. L. The role of tetraether lipid composition in the adaptation of thermophilic archaea to acidity. Front. Microbiol. 4, 62 (2013).
Cooper, J. E. Early interactions between legumes and rhizobia: disclosing complexity in a molecular dialogue. J. Appl. Microbiol. 103, 1355–1365 (2007).
D'Haeze, W. et al. Reactive oxygen species and ethylene play a positive role in lateral root base nodulation of a semiaquatic legume. Proc. Natl Acad. Sci. USA 100, 11789–11794 (2003).
Pauly, N. et al. Reactive oxygen and nitrogen species and glutathione: key players in the legume-Rhizobium symbiosis. J. Exp. Bot. 57, 1769–1776 (2006).
Pierre, O. et al. Peribacteroid space acidification: a marker of mature bacteroid functioning in Medicago truncatula nodules. Plant Cell Environ. 36, 2059–2070 (2013).
Miller, K. J. & Wood, J. M. Osmoadaptation by rhizosphere bacteria. Annu. Rev. Microbiol. 50, 01–36 (1996).
Czernic, P. et al. Convergent evolution of endosymbiont differentiation in Dalbergoid and IRLC legumes mediated by nodule-specific cysteine-rich peptides. Plant Physiol. 169, 1254–1265 (2015).
Haag, A. F. et al. Molecular insights into bacteroid development during Rhizobium-legume symbiosis. FEMS Microbiol. Rev. 37, 364–383 (2013).
Kondorosi, E., Mergaert, P. & Kereszt, A. A paradigm for endosymbiotic life: cell differentiation of Rhizobium bacteria provoked by host plant factors. Annu. Rev. Microbiol. 67, 611–628 (2013).
Sprent, J., Ardley, J. & James, E. Biogeography of nodulated legumes and their nitrogen fixing symbionts. New Phytol. 215, 40–56 (2017).
Parker, M. A. The spread of Bradyrhizobium lineages across host legume clades: from Abarema to Zygia. Microb. Ecol. 69, 630–640 (2015).
Fowler, D. et al. Effects of global change during the 21st century on the nitrogen cycle. Atmos. Chem. Phys. 15, 13849–13893 (2015).
Volkman, J. K. Sterols and other triterpenoids: source specificity and evolution of biosynthetic pathways. Org. Geochem. 36, 139 (2005).
Killops, S. D. & Killops, V. in Introduction to Organic Geochemistry 2nd edn Ch. 2 (Blackwell Publishing, Oxford, 2005).
Sohlenkamp, C. & Geiger, O. Bacterial membrane lipids: Diversity in structures and pathways. FEMS Microbiol. Rev. 40, 133–159 (2015).
Raetz, C. R. & Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700 (2002).
Silipo, A., De Castro, C., Lanzetta, R., Parrilli, M. & Molinaro, A. in Prokaryotic Cell Wall Compounds: Structure and Biochemistry (eds König, H., Claus, H. & Varma, A.) 133–153 (Springer, Berlin and Heidelberg, Germany, 2010).
Molinaro, A. et al. Chemistry of lipid A: at the heart of innate immunity. Chemistry 21, 500–519 (2015).
Róg, T., Pasenkiewicz-Gierula, M., Vattulainen, I. & Karttunen, M. Ordering effects of cholesterol and its analogues. Biochim. Biophys. Acta 1788, 97–121 (2009).
Van Meer, G., Voelker, D. R. & Feigenson, G. W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124 (2008).
Harriott, O. T., Khairallah, L. & Benson, D. R. Isolation and structure of the lipid envelopes from the nitrogen-fixing vesicles of Frankia sp. Strain CpI1. J. Bacteriol. 173, 2061–2067 (1991).
Berry, A. M. et al. Hopanoid lipids compose the Frankia vesicle envelope, presumptive barrier of oxygen diffusion to nitrogenase. Proc. Natl Acad. Sci. USA 90, 6091–6094 (1993). This work identifies hopanoids as the primary component of vesicles in Frankia spp., suggesting that hopanoids limit oxygen diffusion during nitrogen fixation.
Delgado-Baquerizo, M. et al. A global atlas of the dominant bacteria found in soil. Science 359, 320–325 (2018).
Acknowledgements
The authors thank A. Session, P. Normand and the reviewers for constructive comments on the manuscript. We appreciate permission from D. Benson, A. Berry and J. Sáenz to reproduce images from their work. Grants from the Howard Hughes Medical Institute (HHMI; D.K.N.), National Aeronautics and Space Administration (NASA; NNX12AD93G, D.K.N.), the Jane Coffin Childs Memorial Fund (B.J.B.), the US National Institutes of Health (NIH; K99GM126141, B.J.B.), H2020- MSCA-ITN-2014-ETN TOLLerant (A.S.), Progetto Galileo G14-23 (A.S.), Mizutani Foundation for Glycoscience 2014 (A.M.) and the French National Research Agency (ANR-BugsInaCell-13-BSV7-0013) have sustained our research on this problem.
Author information
Authors and Affiliations
Contributions
B.J.B, E.G., A.S. and D.K.N. researched data for the article. B.J.B., E.G., A.M., A.S. and D.K.N. substantially contributed to the discussion of content. B.J.B., N.B., E.G., A.S. and D.K.N. wrote the article. All authors reviewed and edited the manuscript before submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Hopanoids
-
Pentacyclic lipids with C6 A-D rings and a C5 E ring, among which six core methyl groups are distributed. Hopanoids are also characterized by the formation of accessory groups at their C2, C3 and C30 positions.
- Sterols
-
Tetracyclic lipids with C6 A-C rings and a C5 D ring. The parent compounds for all sterols contain oxygen groups at the C3 position.
- Terpenoids
-
Molecules assembled from two or more C5 isoprene units that share the core formula (C5H8)n. They are also known as isoprenoids.
- Triterpenoid
-
Molecules derived from assemblies of six isoprene units with the core formula (C5H8)6 or C30H48. They may be acyclic or cyclic.
- Squalene
-
An acyclic triterpene with an irregular (tail-to-tail) linkage between two 3-isoprene units; a parent molecule of cyclic triterpenoids.
- Triterpenoid cyclase
-
A superfamily of enzymes that convert acyclic triterpenoids, including squalene and 2,3-oxidosqualene, into various cyclic products.
- Oxidosqualene cyclases
-
(OSCs). A family of predominantly eukaryotic 3-β-hydroxytriterpene cyclases that transform 2,3-oxidosqualene into sterols and diverse other cyclic triterpenoids.
- Squalene-hopene cyclases
-
A family of predominantly bacterial 3-deoxytriterpene cyclases that cyclize squalene to primarily form hopanoids.
- C30 hopanoids
-
Short hopanoids containing no additional carbon atoms that are not derived from squalene.
- Tetrahymanol
-
A hopanoid-like compound with a C6 E-ring. In bacteria, it is made from E-ring expansion of the C30 hopanoid diploptene.
- Legume
-
Legumes are flowering plants of the Fabaceae (previously known as Leguminosae) family, which includes economically important crops such as soybeans, common beans and peanuts.
- C35 hopanoids
-
Extended hopanoids that contain ribose-derived hydrocarbon side chains at their C30 position.
- 2-Methyl-hopanoids
-
(2Me-hopanoids). Hopanoids containing an accessory methyl group at the C2 position.
- 3-Methyl-hopanoids
-
(3Me-hopanoids). Hopanoids containing an accessory methyl group at the C3 position.
- Liposomes
-
Spherical vesicles surrounded by one or more lipid bilayers that can be produced from cellular membranes in vivo or synthetically by sonication or extrusion of lipids into aqueous solution.
- Bacteriohopanetetrol
-
(BHT). A common C35 hopanoid containing a tetra-hydroxylated C5 side chain.
- Membrane fluidity
-
The rotational and diffusional freedom of movement of molecules within a membrane.
- Lipid rafts
-
Membrane microdomains with high stability that are thought to recruit specific membrane-associated proteins to spatially regulate their functions.
- Lipopolysaccharides
-
(LPSs). Complex, heat-stable amphiphilic lipids that are the main component of the external leaflet of the outer membrane of Gram-negative bacteria.
- Lipid A
-
The lipophilic moiety of lipopolysaccharides.
- Hypoxic
-
An environment with a low concentration of oxygen (usually less than 30% saturation).
- Nitrogen fixation
-
The conversion of dinitrogen gas into fixed or bioavailable nitrogen sources such as ammonia.
- Nitrogenase
-
A bacterial enzyme complex that performs the following reaction: N2 + 16ATP + 10H+ + 8e− → 2NH4+ + H2 + 16ADP−Pi
- Root nodule
-
A specialized root organ that is generated by most legume plants to house nitrogen-fixing symbionts and create a specialized microenvironment to support bacterial nitrogen fixation.
- Rhizobia
-
A paraphyletic group of nitrogen-fixing soil bacteria that can engage in symbioses with legumes.
- Leghaemoglobin
-
An oxygen-carrying haem protein expressed in the root nodules of rhizobial host plants.
- Aeschynomene
-
A genus of tropical legumes that is broadly distributed globally and is used for livestock grazing.
- Hopanoid-lipid A
-
(HoLa). An extended hopanoid that is covalently attached to lipid A and appears to be unique to the Bradyrhizobiaceae.
Rights and permissions
About this article
Cite this article
Belin, B., Busset, N., Giraud, E. et al. Hopanoid lipids: from membranes to plant–bacteria interactions. Nat Rev Microbiol 16, 304–315 (2018). https://doi.org/10.1038/nrmicro.2017.173
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro.2017.173
This article is cited by
-
Integration of (S)-2,3-oxidosqualene enables E. coli to become Iron Man E. coli with improved overall tolerance
Biotechnology for Biofuels and Bioproducts (2023)
-
Cultivation and genomic characterization of novel and ubiquitous marine nitrite-oxidizing bacteria from the Nitrospirales
The ISME Journal (2023)
-
Genetics re-establish the utility of 2-methylhopanes as cyanobacterial biomarkers before 750 million years ago
Nature Ecology & Evolution (2023)
-
Design of a redox-proficient Escherichia coli for screening terpenoids and modifying cytochrome P450s
Nature Catalysis (2023)
-
Revealing potential functions of hypothetical proteins induced by genistein in the symbiosis island of Bradyrhizobium japonicum commercial strain SEMIA 5079 (= CPAC 15)
BMC Microbiology (2022)