Key Points
-
Apoptosis and autophagy constitute the two self-destructive processes by which supernumerary, damaged or aged cells and organelles are eliminated. Beyond this homeostatic function, autophagy is also a process through which cells adapt their metabolisms to starvation, imposed by decreased extracellular nutrients or by decreased intracellular metabolite concentrations that result from growth-factor signalling.
-
The relationship between autophagy and cell death is complex because autophagy constitutes an adaptive response to different kinds of stress by which the cells avoid cell death; yet, in some settings, it can also contribute to the demise of cells.
-
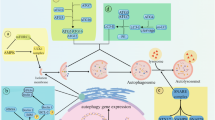
In response to the same panoply of stressors, cells can preferentially undergo apoptosis or autophagy, a choice that is dictated by the intensity of the stimulus and thresholds for either response. Several stress mediators (reactive oxygen species, ceramide, elevation of cytosolic Ca2+), isoforms of p19ARF (or its human homologue, p14ARF), p53, BH3-only proteins and death-associated protein kinases (DAPK family members) can stimulate both apoptosis and autophagy.
-
Organellar stress that affects mitochondria and the endoplasmic reticulum can induce a specific autophagic response that leads to the removal of damaged organelles (mitophagy and reticulophagy, respectively) and protects cells. Beyond a threshold (which is lowered when autophagy is inhibited), such stress causes apoptosis.
-
Several proteins that have an essential role in autophagy have a direct or indirect impact on the regulation or execution of apoptosis. As an example, Atg5 is an essential autophagy inducer, yet it can be cleaved by calpain cysteine proteases to lose its pro-autophagy effects and become a pro-apoptotic molecule.
-
Beclin-1 (or its yeast homologue Atg6) interacts with the anti-apoptotic multidomain proteins of the BCL2 family (in particular BCL2 and BCL-XL) through a BH3 domain. BH3-only proteins and pharmacological BH3 mimetics competitively disrupt the inhibitory interaction between beclin-1 and BCL2 or BCL-XL, thereby stimulating autophagy. Thus, BH3 domains, which are well known for their apoptosis-inducing property, can also stimulate autophagy.
-
The pharmacological stimulation of autophagy may have cytoprotective effects, for example, in mouse models of neurodegenerative disease, whereas the inhibition of autophagy can sensitize cancer cells to chemotherapy to promote p53-induced apoptosis. Thus, manipulation of autophagy can determine cell-fate decisions in clinical settings.
Abstract
The functional relationship between apoptosis ('self-killing') and autophagy ('self-eating') is complex in the sense that, under certain circumstances, autophagy constitutes a stress adaptation that avoids cell death (and suppresses apoptosis), whereas in other cellular settings, it constitutes an alternative cell-death pathway. Autophagy and apoptosis may be triggered by common upstream signals, and sometimes this results in combined autophagy and apoptosis; in other instances, the cell switches between the two responses in a mutually exclusive manner. On a molecular level, this means that the apoptotic and autophagic response machineries share common pathways that either link or polarize the cellular responses.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Danial, N. N. & Korsmeyer, S. J. Cell death: critical control points. Cell 116, 205–219 (2004).
Green, D. R. Apoptotic pathways: ten minutes to dead. Cell 121, 671–674 (2005).
Kroemer, G. et al. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death. Cell Death Differ. 12 (Suppl. 2), 1463–1467 (2005).
Shintani, T. & Klionsky, D. J. Autophagy in health and disease: a double-edged sword. Science 306, 990–995 (2004).
Rubinsztein, D. C., Gestwicki, J. E., Murphy, L. O. & Klionsky, D. J. Potential therapeutic applications of autophagy. Nature Rev. Drug Discov. 6, 304–312 (2007). A landmark review that enumerates the potential therapeutic possibilities of autophagy inhibition and stimulation.
Lum, J. J., DeBerardinis, R. J. & Thompson, C. B. Autophagy in metazoans: cell survival in the land of plenty. Nature Rev. Mol. Cell Biol. 6, 439–448 (2005).
Baehrecke, E. H. Autophagy: dual roles in life and death? Nature Rev. Mol. Cell Biol. 6, 505–510 (2005).
Levine, B. & Yuan, J. Autophagy in cell death: an innocent convict? J. Clin. Invest. 115, 2679–2688 (2005).
Kroemer, G. & Jaattela, M. Lysosomes and autophagy in cell death control. Nature Rev. Cancer 5, 886–897 (2005).
Kondo, Y., Kanzawa, T., Sawaya, R. & Kondo, S. The role of autophagy in cancer development and response to therapy. Nature Rev. Cancer 5, 726–734 (2005).
Gozuacik, D. & Kimchi, A. Autophagy and cell death. Curr. Top. Dev. Biol. 78, 217–245 (2007).
Lockshin, R. A. & Zakeri, Z. Programmed cell death and apoptosis: origins of the theory. Nature Rev. Mol. Cell Biol. 2, 545–550 (2001). A fascinating historical overview on the theory of developmental and homeostatic cell death.
Shimizu, S. et al. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nature Cell Biol. 6, 1221–1228 (2004).
Madden, D. T., Egger, L. & Bredesen, D. E. A calpain-like protease inhibits autophagic cell death. Autophagy 3, 519–522 (2007).
Xu, Y., Kim, S. O., Li, Y. & Han, J. Autophagy contributes to caspase-independent macrophage cell death. J. Biol. Chem. 281, 19179–19187 (2006).
Yu, L. et al. Regulation of an ATG7–beclin 1 program of autophagic cell death by caspase-8. Science 304, 1500–1502 (2004).
Yu, L. et al. Autophagic programmed cell death by selective catalase degradation. Proc. Natl Acad. Sci. USA 103, 4952–4957 (2006). Provides the first example of how selective degradation of a vital cellular protein by autophagy can precipitate cell death.
Lum, J. J. et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell 120, 237–248 (2005). Reports the discovery of a close link between autophagy, cellular atrophy and the absence of growth-factor signalling in which autophagy allows cells to adapt to a failing supply of metabolites.
Gonzalez-Polo, R. A. et al. The apoptosis/autophagy paradox: autophagic vacuolization before apoptotic death. J. Cell Sci. 118, 3091–3102 (2005).
Boya, P. et al. Inhibition of macroautophagy triggers apoptosis. Mol. Cell Biol. 25, 1025–1040 (2005). The first report showing that autophagy avoids apoptosis in the context of growth-factor and nutrient depletion.
Hara, T. et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889 (2006).
Komatsu, M. et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441, 880–884 (2006).
Pua, H. H., Dzhagalov, I., Chuck, M., Mizushima, N. & He, Y. W. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J. Exp. Med. 204, 25–31 (2007).
Takacs-Vellai, K. et al. Inactivation of the autophagy gene bec-1 triggers apoptotic cell death in C. elegans. Curr. Biol. 15, 1513–1517 (2005).
Nutt, L. K. et al. Metabolic regulation of oocyte cell death through the CaMKII-mediated phosphorylation of caspase-2. Cell 123, 89–103 (2005).
Kroemer, G., Galluzzi, L. & Brenner, C. Mitochondrial membrane permeabilization in cell death. Physiol. Rev. 87, 99–163 (2007).
Golstein, P. & Kroemer, G. Cell death by necrosis: towards a molecular definition. Trends Biochem. Sci. 32, 37–43 (2007).
Katayama, M., Kawaguchi, T., Berger, M. S. & Pieper, R. O. DNA damaging agent-induced autophagy produces a cytoprotective adenosine triphosphate surge in malignant glioma cells. Cell Death Differ. 14, 548–558 (2007).
Degenhardt, K. et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 10, 51–64 (2006). A fascinating study of the potential role of autophagy in the growth of cancer cells in vivo.
Williams, A. et al. Aggregate-prone proteins are cleared from the cytosol by autophagy: therapeutic implications. Curr. Top. Dev. Biol. 76, 89–101 (2006).
Ravikumar, B. et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nature Genet. 36, 585–595 (2004). The first report to show that pharmacological stimulation of autophagy may reduce the histopathological and clinical signs of neurodegeneration in vivo.
Sarkar, S. et al. Lithium induces autophagy by inhibiting inositol monophosphatase. J. Cell Biol. 170, 1101–1111 (2005).
Shibata, M. et al. Regulation of intracellular accumulation of mutant huntingtin by beclin 1. J. Biol. Chem. 281, 14474–14485 (2006).
Cuervo, A. M. Autophagy in neurons: it is not all about food. Trends Mol. Med. 12, 461–464 (2006).
Ravikumar, B., Berger, Z., Vacher, C., O'Kane, C. J. & Rubinsztein, D. C. Rapamycin pre-treatment protects against apoptosis. Hum. Mol. Genet. 15, 1209–1216 (2006).
Colell, A. et al. GAPDH and autophagy preserve cellular survival during caspase-independent cell death. Cell 129, 983–997 (2007). A highly intriguing report suggesting that GAPDH can stimulate autophagy by its action as a nuclear transcription factor, and thereby facilitates the survival of cells that have undergone MOMP.
Liang, J. et al. The energy sensing LKB1–AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nature Cell Biol. 9, 218–224 (2007).
Clarke, P. G. Developmental cell death: morphological diversity and multiple mechanisms. Anat. Embryol. (Berl.) 181, 195–213 (1990).
Martin, D. N. & Baehrecke, E. H. Caspases function in autophagic programmed cell death in Drosophila. Development 131, 275–284 (2004).
Scott, R. C., Juhasz, G. & Neufeld, T. P. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr. Biol. 17, 1–11 (2007). Reports an in vivo experiment that shows how autophagy can indirectly trigger cell death through the induction of apoptosis.
Espert, L. et al. Autophagy is involved in T cell death after binding of HIV-1 envelope proteins to CXCR4. J. Clin. Invest. 116, 2161–2172 (2006).
Yue, Z., Jin, S., Yang, C., Levine, A. J. & Heintz, N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl Acad. Sci. USA 100, 15077–15082 (2003).
Qu, X. et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 112, 1809–1820 (2003).
Mathew, R. et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 21, 1367–1381 (2007). This excellent paper suggests a mechanistic explanation for the tumour-suppressive function of beclin-1, which is required for genomic stability.
Scherz-Shouval, R. et al. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 26, 1749–1760 (2007).
Lavieu, G. et al. Regulation of autophagy by sphingosine kinase 1 and its role in cell survival during nutrient starvation. J. Biol. Chem. 281, 8518–8527 (2006).
Hait, N. C., Oskeritzian, C. A., Paugh, S. W., Milstien, S. & Spiegel, S. Sphingosine kinases, sphingosine 1-phosphate, apoptosis and diseases. Biochim. Biophys. Acta 1758, 2016–2026 (2006).
Rizzuto, R. & Pozzan, T. Microdomains of intracellular Ca2+: molecular determinants and functional consequences. Physiol. Rev. 86, 369–408 (2006).
Hoyer-Hansen, M. et al. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-β, and Bcl-2. Mol. Cell 25, 193–205 (2007).
Demarchi, F. et al. Calpain is required for macroautophagy in mammalian cells. J. Cell Biol. 175, 595–605 (2006).
Yousefi, S. et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nature Cell Biol. 8, 1124–1132 (2006). An intriguing report that documents how ATG5 protein can switch from its normal function in autophagy to a pro-apoptotic one.
Vousden, K. H. & Lane, D. P. p53 in health and disease. Nature Rev. Mol. Cell Biol. 8, 275–283 (2007).
Zeng, X., Yan, T., Schupp, J. E., Seo, Y. & Kinsella, T. J. DNA mismatch repair initiates 6-thioguanine-induced autophagy through p53 activation in human tumor cells. Clin. Cancer Res. 13, 1315–1321 (2007).
Feng, Z., Zhang, H., Levine, A. J. & Jin, S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc. Natl Acad. Sci. USA 102, 8204–8209 (2005).
Crighton, D. et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 126, 121–134 (2006).
Galonek, H. L. & Hardwick, J. M. Upgrading the BCL-2 network. Nature Cell Biol. 8, 1317–1319 (2006).
Adams, J. M. & Cory, S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene 26, 1324–1337 (2007).
Maiuri, M. C. et al. Functional and physical interaction between Bcl-XL and a BH3-like domain in Beclin-1. EMBO J. 26, 2527–2539 (2007). This paper explains the triangular relationship between the essential autophagy protein beclin-1, the anti-apoptotic multidomain proteins of the BCL2 family and BH3-only proteins.
Kessel, D. & Reiners, J. J. Jr. Initiation of apoptosis and autophagy by the Bcl-2 antagonist HA14–1. Cancer Lett. 249, 294–299 (2007).
Daido, S. et al. Pivotal role of the cell death factor BNIP3 in ceramide-induced autophagic cell death in malignant glioma cells. Cancer Res. 64, 4286–4293 (2004).
Hamacher-Brady, A. et al. Response to myocardial ischemia/reperfusion injury involves Bnip3 and autophagy. Cell Death Differ. 14, 146–157 (2007).
Oberstein, A., Jeffrey, P. & Shi, Y. Crystal structure of the BCL-XL-beclin 1 peptide complex: beclin 1 is a novel BH3-only protein. J. Biol. Chem. 282, 13123–13132 (2007).
Maiuri, M. C. et al. BH3-only proteins and BH3 mimetics induce autophagy by competitively disrupting the interaction between beclin 1 and Bcl-2/Bcl-XL . Autophagy 3, 374–376 (2007).
Nobukuni, T., Kozma, S. C. & Thomas, G. hvps34, an ancient player, enters a growing game: mTOR complex1/S6K1 signaling. Curr. Opin. Cell Biol. 19, 135–141 (2007).
Bialik, S. & Kimchi, A. The death-associated protein kinases: structure, function, and beyond. Annu. Rev. Biochem. 75, 189–210 (2006).
Raveh, T., Droguett, G., Horwitz, M. S., DePinho, R. A. & Kimchi, A. DAP kinase activates a p19ARF/p53-mediated apoptotic checkpoint to suppress oncogenic transformation. Nature Cell Biol. 3, 1–7 (2001).
Wang, W. J., Kuo, J. C., Yao, C. C. & Chen, R. H. DAP-kinase induces apoptosis by suppressing integrin activity and disrupting matrix survival signals. J. Cell Biol. 159, 169–179 (2002).
Jang, C. W. et al. TGF-β induces apoptosis through Smad-mediated expression of DAP-kinase. Nature Cell Biol. 4, 51–58 (2002).
Inbal, B., Bialik, S., Sabanay, I., Shani, G. & Kimchi, A. DAP kinase and DRP-1 mediate membrane blebbing and the formation of autophagic vesicles during programmed cell death. J. Cell Biol. 157, 455–468 (2002).
Shani, G. et al. Death-associated protein kinase phosphorylates ZIP kinase, forming a unique kinase hierarchy to activate its cell death functions. Mol. Cell Biol. 24, 8611–8626 (2004).
Raval, A. et al. Downregulation of death-associated protein kinase 1 (DAPK1) in chronic lymphocytic leukemia. Cell 129, 879–890 (2007)
Reef, S. et al. A short mitochondrial form of p19ARF induces autophagy and caspase-independent cell death. Mol. Cell 22, 463–475 (2006). Demonstrates that a short isoform of the ARF protein (generated by internal initiation of translation) can induce autophagy, presumably through an action on depolarized but not permeabilized mitochondria.
Rodriguez-Enriquez, S., Kim, I., Currin, R. T. & Lemasters, J. J. Tracker dyes to probe mitochondrial autophagy (mitophagy) in rat hepatocytes. Autophagy 2, 39–46 (2006).
Vande Velde, C. et al. BNIP3 and genetic control of necrosis-like cell death through the mitochondrial permeability transition pore. Mol. Cell Biol. 20, 5454–5468 (2000).
Zhu, J. H. et al. Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridinium-induced cell death. Am. J. Pathol. 170, 75–86 (2007).
Xu, C., Bailly-Maitre, B. & Reed, J. C. Endoplasmic reticulum stress: cell life and death decisions. J. Clin. Invest. 115, 2656–2664 (2005).
Yorimitsu, T., Nair, U., Yang, Z. & Klionsky, D. J. Endoplasmic reticulum stress triggers autophagy. J. Biol. Chem. 281, 30299–30304 (2006).
Schroder, M. & Kaufman, R. J. The mammalian unfolded protein response. Annu. Rev. Biochem. 74, 739–789 (2005).
Criollo, A. et al. Regulation of autophagy by the inositol trisphosphate receptor. Cell Death Differ. 14, 1029–1039 (2007).
Hetz, C. et al. Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1α. Science 312, 572–576 (2006).
Boya, P., Cohen, I., Zamzami, N., Vieira, H. L. & Kroemer, G. Endoplasmic reticulum stress-induced cell death requires mitochondrial membrane permeabilization. Cell Death Differ. 9, 465–467 (2002).
Ogata, M. et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol. Cell Biol. 26, 9220–9231 (2006). Shows for the first time that ER stress can stimulate autophagy in mammalian cells.
Kouroku, Y. et al. ER stress (PERK/eIF2α phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 14, 230–239 (2007).
Ding, W. X. et al. Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J. Biol. Chem. (2006).
Pyo, J. O. et al. Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J. Biol. Chem. 280, 20722–20729 (2005).
Liang, C. et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nature Cell Biol. 8, 688–699 (2006).
Fimia, G. M. et al. Ambra1 regulates autophagy and development of the nervous system. Nature 24 Jun 2007 (doi:10.1038/nature05925). References 85 and 87 report the identification of two beclin-1-binding proteins that are required for the induction of autophagy.
Pattingre, S. et al. Bcl-2 antiapoptotic proteins inhibit beclin 1-dependent autophagy. Cell 122, 927–939 (2005).
Yanagisawa, H., Miyashita, T., Nakano, Y. & Yamamoto, D. HSpin1, a transmembrane protein interacting with Bcl-2/Bcl-XL, induces a caspase-independent autophagic cell death. Cell Death Differ. 10, 798–807 (2003).
Qu, X. et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell 128, 931–946 (2007). Indicates that, during developmental cell death, autophagy may be a mechanism that occurs before (but independently of) apoptosis and allows for optimal heterophagic removal of the dying cell.
Tanida, I., Ueno, T. & Kominami, E. LC3 conjugation system in mammalian autophagy. Int. J. Biochem. Cell Biol. 36, 2503–2518 (2004).
Levine, B. & Klionsky, D. J. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6, 463–477 (2004).
Krammer, P. H. CD95's deadly mission in the immune system. Nature 407, 789–795 (2000).
Tinel, A. et al. Autoproteolysis of PIDD marks the bifurcation between pro-death caspase-2 and pro-survival NF-κB pathway. EMBO J. 26, 197–208 (2007).
Kroemer, G. & Martin, S. J. Caspase-independent cell death. Nature Med. 11, 725–730 (2005).
Arama, E., Agapite, J. & Steller, H. Caspase activity and a specific cytochrome c are required for sperm differentiation in Drosophila. Dev. Cell 4, 687–697 (2003).
Kuranaga, E. & Miura, M. Nonapoptotic functions of caspases: caspases as regulatory molecules for immunity and cell-fate determination. Trends Cell Biol. 17, 135–144 (2007).
Jonas, E. BCL-XL regulates synaptic plasticity. Mol. Interv. 6, 208–222 (2006).
Zermati, Y. et al. Non-apoptotic role for Apaf-1 in the DNA damage response. Mol. Cell in the press.
Acknowledgements
The authors are supported by grants from the Ligue Nationale contre le Cancer (équipe labellisée), the European Union (Death-Train, ChemoRes, Trans-Death, Right, active p53), Cancéropôle Ile-de-France, Institut National du Cancer, Agence Nationale pour la Recherche (to G.K.) and the Center of Excellence grant from the Flight Attendant Medical Research Institute (FAMRI) (to A.K.). A.K. is the incumbent Helena Rubinstein Chair of Cancer Research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information S1 (table)
Examples of cell death exacerbated by inhibition of autophagy (PDF 159 kb)
Related links
Related links
DATABASES
OMIM
FURTHER INFORMATION
Glossary
- Autophagy
-
In this context, autophagy is used synonymously with macroautophagy, a process in which a portion of the cytoplasm is engulfed by a specific membrane and later digested by lysosomal enzymes.
- Programmed cell death
-
A genetically controlled cell-death process that is turned on in response to external or internal signals.
- Lysosome
-
A membrane-surrounded sac that contains >50 acid hydrolases (phosphatases, nucleases, glycosidases, proteases, peptidases, sulphatases and lipases), which can digest all of the major macromolecules of the cell to breakdown products that are then available for metabolic reuse.
- Catabolism
-
A metabolic state that leads to the degradation of macromolecules to yield small molecules and/or ATP, usually as a consequence of nutrient deprivation. Catabolic reactions include glycogenolysis, β-oxidation of fatty acids and autophagy.
- BCL2 family
-
A family of proteins that contain at least one BCL2 homology (BH) region. The family is divided into anti-apoptotic multidomain proteins (such as BCL2, BCL-XL and MCL1), which contain four BH domains (BH1, BH2, BH3, BH4), pro-apoptotic multidomain proteins (for example, BAX and BAK), which contain BH1, BH2 and BH3, and the pro-apoptotic BH3-only protein family.
- Mitochondrial outer membrane permeabilization
-
(MOMP). An apoptosis-associated process that results in apoptosis-inducing proteins (such as cytochrome c, apoptosis-inducing factor, Smac/Diablo) that are normally retained in the mitochondrial intermembrane space being released through the outer membrane into the cytosol.
- Autolysosome
-
Also called autophagolysosome. A vesicle that is formed by the fusion of an autophagosome (or an amphisome) with a lysosome.
- Caspase
-
One of a family of cysteine proteases that cleave after aspartate residues in substrate proteins. Initiator caspases are typically activated in response to a specific triggering event (for example, caspase-8 upon death-receptor ligation, caspase-9 upon apoptosome activation, caspase-2 upon DNA damage), whereas effector caspases (mainly caspase-3, -6 and -7) are important for the ordered dismantling of vital cellular structures.
- Reactive oxygen species
-
(ROS). Reduced derivatives of molecular oxygen (O2), including classical oxygen radicals and peroxides, which are formed within cells.
- Death receptor
-
One of a family of cell-surface receptors that mediate cell death upon ligand-induced trimerization. The best-studied members include tumour necrosis factor receptor-1 (TNFR1), FAS (or CD95, which binds FAS ligand) and two receptors for TNF-related apoptosis-inducing ligand (TRAILR1 and -R2).
- Cytochrome c
-
A haem protein that is normally confined to the mitochondrial intermembrane space. Upon induction of apoptosis, cytochrome c is released from mitochondria and triggers the formation of the apoptosome, a caspase activation complex.
- Apoptosome
-
A complex that forms when cytochrome c is released from mitochondria and interacts with the cytosolic protein APAF1, which, in turn, recruits pro-caspase-9. In the presence of ATP, this interaction results in the allosteric activation of caspase-9 and in the subsequent activation of the effector caspase-3.
- Apoptosis-inducing factor
-
(AIF). A flavoprotein that is normally present in the mitochondrial intermembrane space. Following apoptosis induction, AIF translocates to the nucleus where it activates a molecular complex that causes large-scale DNA fragmentation, presumably in a caspase-independent fashion.
- Lysosomal membrane permeabilization
-
(LMP). A perturbation of lysosomal membrane function, leading to the translocation of lysosomal hydrolases (including cathepsins) from the lysosomal lumen to the rest of the cell. LMP can be induced by endogenous signal transducers (such as reactive oxygen species and sphingosine) as well as by lysosomotropic drugs.
- Cathepsin
-
A protease that localizes mainly to lysosomes and lysosome-like organelles. Cathepsin proteases can be divided into three subgroups according to their active-site amino acid: cysteine (cathepsins B, C, H, F, K, L, O, S, V, W and X/Z), aspartate (cathepsins D and E) and serine (cathepsin G).
- Haploinsufficient
-
A gene that requires biallelic expression to produce the amount of protein that is sufficient to guarantee a normal biological function.
- Tumour-suppressor gene
-
A gene that, when eliminated or inactivated, is permissive for the development of cancers. These genes often determine cell-cycle checkpoints or facilitate the induction of programmed cell death.
- p53
-
A transcription factor that is activated by numerous genotoxic insults to induce cell-cycle arrest, cellular senescence or apoptosis. p53 is frequently mutated or functionally inactivated in cancer.
- Oncogene
-
A gene of which overexpression or gain-of-function mutation contributes to oncogenesis.
- Unfolded protein response
-
(UPR). A signalling pathway that is activated in response to stress in the endoplasmic reticulum (ER). The UPR can cope effectively with stress by reducing the amount of misfolded protein overload in the ER.
- Heterophagy
-
Phagocytosis of a cell by another cell. Heterophagy has an important role in the efficient removal of apoptotic corpses. Efficient heterophagy is indispensable for avoiding inflammatory responses that are triggered by apoptotic material.
Rights and permissions
About this article
Cite this article
Maiuri, M., Zalckvar, E., Kimchi, A. et al. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol 8, 741–752 (2007). https://doi.org/10.1038/nrm2239
Issue Date:
DOI: https://doi.org/10.1038/nrm2239
This article is cited by
-
The mitochondrial quality control system: a new target for exercise therapeutic intervention in the treatment of brain insulin resistance-induced neurodegeneration in obesity
International Journal of Obesity (2024)
-
Angiogenesis-on-a-chip coupled with single-cell RNA sequencing reveals spatially differential activations of autophagy along angiogenic sprouts
Nature Communications (2024)
-
Autophagy in an extruded disc compared to the remaining disc after lumbar disc herniation in the same patient
European Spine Journal (2024)
-
Mitochondria in Mesenchymal Stem Cells: Key to Fate Determination and Therapeutic Potential
Stem Cell Reviews and Reports (2024)
-
Effects of transport stress on the oxidative index, apoptosis and autophagy in the small intestine of caprine
BMC Veterinary Research (2023)