Key Points
-
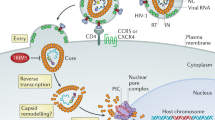
At least two human DNA-cytosine deaminases, APOBEC3F and APOBEC3G, are capable of inhibiting the infection of retroviruses such as HIV-1. This occurs by the high-frequency conversion of retroviral cDNA (minus strand) cytosines to uracils, lesions that, when replicated, result in lethal retroviral plus-strand G to A hypermutations.
-
HIV-1 and many other vertebrate lentiviruses have a counterdefence protein known as Vif (virion infectivity factor), which is able to mediate the proteasomal destruction of these APOBEC3 proteins.
-
HIV-1 Vif is not always effective, as retroviral plus-strand G to A hypermutations are commonly found in patient-derived samples.
-
These observations indicate that the in vivo balance between HIV-1 Vif and the APOBEC3 proteins might be amenable to therapeutic intervention.
-
At least nine other APOBEC3-related cytosine deaminases are encoded by the human genome, and only two of these have well-known functions. APOBEC1 edits C6666 in APOB mRNA, which results in a premature stop codon and a novel protein, whereas AID uses DNA-cytosine deamination to trigger three types of immunoglobulin-gene diversification — somatic hypermutation, gene conversion and class-switch recombination. Parallels between the innate APOBEC3-dependent restriction mechanism and the adaptive AID-dependent antibody response are striking and will probably contribute to future advances in this field.
-
Many of the other APOBEC-family cytosine deaminases probably have as-yet-unappreciated physiological roles. They might target DNA and/or RNA substrates and have important innate immune functions.
-
An examination of other vertebrate genomes shows that the APOBEC3 proteins are specific to mammals and have undergone a relatively recent evolutionary expansion (rodents have one, whereas humans and chimpanzees have eight APOBEC3 genes). We speculate that a protective role in reproduction might constitute a significant part of the selective pressure that drove this gene expansion.
Abstract
A powerful mechanism of vertebrate innate immunity has been discovered in the past year, in which APOBEC proteins inhibit retroviruses by deaminating cytosine residues in nascent retroviral cDNA. To thwart this cellular defence, HIV encodes Vif, a small protein that mediates APOBEC degradation. Therefore, the balance between APOBECs and Vif might be a crucial determinant of the outcome of retroviral infection. Vertebrates have up to 11 different APOBEC proteins, with primates having the most. APOBEC proteins include AID, a probable DNA mutator that is responsible for immunoglobulin-gene diversification, and APOBEC1, an RNA editor with antiretroviral activities. This APOBEC abundance might help to tip the balance in favour of cellular defences.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Dobzhansky, T. Nothing in biology makes sense except in the light of evolution. Am. Biol. Teach. 35, 125–129 (1973).
Beutler, B. & Hoffmann, J. Innate immunity. Curr. Opin. Immunol. 16, 1–3 (2004).
Samuel, C. E. Antiviral actions of interferons. Clin. Microbiol. Rev. 14, 778–809 (2001).
Honjo, T., Muramatsu, M. & Fagarasan, S. AID: how does it aid antibody diversity? Immunity 20, 659–668 (2004).
Neuberger, M. S. et al. Immunity through DNA deamination. Trends Biochem. Sci. 28, 305–312 (2003).
Barre-Sinoussi, F. et al. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 220, 868–871 (1983).
Gallo, R. C. Historical essay. The early years of HIV/AIDS. Science 298, 1728–1730 (2002).
Montagnier, L. Historical essay. A history of HIV discovery. Science 298, 1727–1728 (2002).
Sheehy, A. M. et al. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418, 646–650 (2002). A landmark study identifying human APOBEC3G as a potent inhibitor of Vif-deficient HIV-1 replication and as a functional target of the HIV-1 Vif protein.
Jarmuz, A. et al. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79, 285–296 (2002).
Teng, B., Burant, C. F. & Davidson, N. O. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science 260, 1816–1819 (1993).
Petersen-Mahrt, S. K., Harris, R. S. & Neuberger, M. S. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature 418, 99–103 (2002). This paper proposed the DNA-deamination model for AID-mediated immunoglobin-gene diversification and showed that AID can trigger C/G to T/A transition mutations through a uracil intermediate.
Harris, R. S., Petersen-Mahrt, S. K. & Neuberger, M. S. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol. Cell 10, 1247–1253 (2002). This paper was the first to show that APOBEC3G was capable of DNA cytosine deamination.
Pathak, V. K. & Temin, H. M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proc. Natl Acad. Sci. USA 87, 6019–6023 (1990).
Janini, M. et al. Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4+ T cells. J. Virol. 75, 7973–7986 (2001). This paper provided a systematic analysis and discussion of HIV-1 G to A hypermutation in patient-derived blood cells. The HIV-1 G to A hypermutations occurred exclusively in GA and GG dinucleotide contexts, with GA predominating.
Harris, R. S. et al. DNA deamination mediates innate immunity to retroviral infection. Cell 113, 803–809 (2003).
Lecossier, D. et al. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300, 1112 (2003).
Mangeat, B. et al. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424, 99–103 (2003).
Zhang, H. et al. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424, 94–98 (2003). References 16–19 showed that APOBEC3G is a potent retroviral cDNA deaminase, capable of triggering high levels of retroviral hypermutation. Reference 16 was also the first to report the single-strand-specific DNA-cytosine-deaminase activity of APOBEC3G in vitro.
Beale, R. C. et al. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J. Mol. Biol. 337, 585–596 (2004).
Yu, Q. et al. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nature Struct. Mol. Biol. 11, 435–442 (2004).
Betts, L. et al. Cytidine deaminase. The 2.3 Å crystal structure of an enzyme: transition-state analog complex. J. Mol. Biol. 235, 635–656 (1994).
Johansson, E. et al. Crystal structure of the tetrameric cytidine deaminase from Bacillus subtilis at 2.0 Å resolution. Biochemistry 41, 2563–2570 (2002).
Ko, T. P. et al. Crystal structure of yeast cytosine deaminase. Insights into enzyme mechanism and evolution. J. Biol. Chem. 278, 19111–19117 (2003).
Xie, K. et al. The structure of a yeast RNA-editing deaminase provides insight into the fold and function of activation-induced deaminase and APOBEC-1. Proc. Natl Acad. Sci. USA 101, 8114–8119 (2004).
Shindo, K. et al. The enzymatic activity of CEM15/Apobec-3G is essential for the regulation of the infectivity of HIV-1 virion but not a sole determinant of its antiviral activity. J. Biol. Chem. 278, 44412–44416 (2003).
MacGinnitie, A. J., Anant, S. & Davidson, N. O. Mutagenesis of apobec-1, the catalytic subunit of the mammalian apolipoprotein B mRNA editing enzyme, reveals distinct domains that mediate cytosine nucleoside deaminase, RNA binding, and RNA editing activity. J. Biol. Chem. 270, 14768–14775 (1995).
Navaratnam, N. et al. Evolutionary origins of apoB mRNA editing: catalysis by a cytidine deaminase that has acquired a novel RNA-binding motif at its active site. Cell 81, 187–195 (1995).
Navaratnam, N. et al. Escherichia coli cytidine deaminase provides a molecular model for apoB RNA editing and a mechanism for RNA substrate recognition. J. Mol. Biol. 275, 695–714 (1998).
Ta, V. T. et al. AID mutant analyses indicate requirement for class-switch-specific cofactors. Nature Immunol. 4, 843–848 (2003).
Svarovskaia, E. S. et al. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J. Biol. Chem. 279, 35822–35828 (2004). This paper and references 36 and 37 are beginning to shed light on the intriguing mechanism of how APOBEC3G is incorporated into the HIV-1 virion, through Gag, RNA and/or a Gag–RNA complex.
Stopak, K. et al. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12, 591–601 (2003).
Marin, M. et al. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nature Med. 9, 1398–1403 (2003).
Lellek, H. et al. Purification and molecular cloning of a novel essential component of the apolipoprotein B mRNA editing enzyme-complex. J. Biol. Chem. 275, 19848–19856 (2000).
Mehta, A. et al. Molecular cloning of Apobec-1 complementation factor, a novel RNA-binding protein involved in the editing of apolipoprotein B mRNA. Mol. Cell. Biol. 20, 1846–1854 (2000).
Alce, T. M. & Popik, W. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J. Biol. Chem. 279, 34083–34086 (2004).
Cen, S. et al. The interaction between HIV-1 Gag and APOBEC3G. J. Biol. Chem. 279, 33177–33184 (2004).
Li, J., Potash, M. J. & Volsky, D. J. Functional domains of APOBEC3G required for antiviral activity. J. Cell. Biochem. 92, 560–572 (2004).
Berkowitz, R. D. et al. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J. Virol. 69, 6445–6456 (1995).
Mariani, R. et al. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114, 21–31 (2003).
Bishop, K. N. et al. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14, 1392–1396 (2004).
Wiegand, H. L. et al. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23, 2451–2458 (2004).
Zheng, Y. H. et al. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 78, 6073–6076 (2004). References 41–43 and 58 have shown that some other APOBEC-family members (in addition to APOBEC3G) can also function to restrict retroviral infection. An important implication is that APOBEC3F might be the dominant restrictor of HIV-1 infection in vivo , with APOBEC3G having a key supporting role.
Kobayashi, M. et al. APOBEC3G targets specific virus species. J. Virol. 78, 8238–8244 (2004).
Gabuzda, D. H. et al. Role of Vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J. Virol. 66, 6489–6495 (1992).
von Schwedler, U. et al. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J. Virol. 67, 4945–4955 (1993).
Conticello, S. G., Harris, R. S. & Neuberger, M. S. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 13, 2009–2013 (2003).
Sheehy, A. M., Gaddis, N. C. & Malim, M. H. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nature Med. 9, 1404–1407 (2003).
Yu, X. et al. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif–Cul5–SCF complex. Science 302, 1056–1060 (2003). This paper revealed that HIV-1 Vif functions to recruit a ubiquitin-ligase complex to accomplish the degradation of APOBEC3G.
Kao, S. et al. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J. Virol. 77, 11398–11407 (2003).
Bogerd, H. P. et al. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc. Natl Acad. Sci. USA 101, 3770–3774 (2004).
Mangeat, B. et al. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J. Biol. Chem. 279, 14481–14483 (2004).
Schrofelbauer, B., Chen, D. & Landau, N. R. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc. Natl Acad. Sci. USA 101, 3927–3932 (2004).
Xu, H. et al. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc. Natl Acad. Sci. USA 101, 5652–5657 (2004).
Schrofelbauer, B., Yu, Q. & Landau, N. R. New insights into the role of Vif in HIV-1 replication. AIDS Rev. 6, 34–39 (2004).
Navarro, F. & Landau, N. R. Recent insights into HIV-1 Vif. Curr. Opin. Immunol. 16, 477–482 (2004).
Rose, K. M. et al. The viral infectivity factor (Vif) of HIV-1 unveiled. Trends Mol. Med. 10, 291–297 (2004).
Liddament, M. T. et al. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 14, 1385–1391 (2004).
Vartanian, J. P., Sommer, P. & Wain-Hobson, S. Death and the retrovirus. Trends Mol. Med. 9, 409–413 (2003).
Turelli, P. et al. Response to comment on 'Inhibition of hepatitis B virus replication by APOBEC3G'. Science 305, 1403B (2004).
Rosler, C. et al. Comment on 'Inhibition of hepatitis B virus replication by APOBEC3G'. Science 305, 1403; author reply 1403 (2004).
Turelli, P. et al. Inhibition of hepatitis B virus replication by APOBEC3G. Science 303, 1829 (2004). References 60–62 show that APOBEC3G can inhibit replication of HBV, possibly through both deamination-dependent and -independent mechanisms.
Muto, T. et al. Isolation, tissue distribution, and chromosomal localization of the human activation-induced cytidine deaminase (AID) gene. Genomics 68, 85–88 (2000).
Espinosa, R. et al. Assignment of the gene encoding the human apolipoprotein B mRNA editing enzyme (APOBEC1) to chromosome 12p13.1. Genomics 24, 414–415 (1994).
Anant, S. et al. ARCD-1, an apobec-1-related cytidine deaminase, exerts a dominant negative effect on C to U RNA editing. Am. J. Physiol. Cell Physiol. 281, C1904–C1916 (2001).
Liao, W. et al. APOBEC-2, a cardiac- and skeletal muscle-specific member of the cytidine deaminase supergene family. Biochem. Biophys. Res. Commun. 260, 398–404 (1999).
Pham, P. et al. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature 424, 103–107 (2003).
Bishop, K. N. et al. APOBEC-mediated editing of viral RNA. Science 305, 645 (2004). This paper has opened the door to the possibility that APOBEC proteins might also function to restrict infection by RNA viruses. It showed that rat APOBEC1 can edit HIV-1 RNA, as well as deaminate its cDNA.
Ramiro, A. R. et al. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nature Immunol. 4, 452–456 (2003).
Wedekind, J. E. et al. Messenger RNA editing in mammals: new members of the APOBEC family seeking roles in the family business. Trends Genet. 19, 207–216 (2003).
Zhang, J. & Webb, D. M. Rapid evolution of primate antiviral enzyme APOBEC3G. Hum. Mol. Genet. 13, 1785–1791 (2004).
Sawyer, S. L., Emerman, M. & Malik, H. S. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2, e275 (2004).
Harris, R. S. et al. AID is essential for immunoglobulin V gene conversion in a cultured B cell line. Curr. Biol. 12, 435–438 (2002).
Arakawa, H., Hauschild, J. & Buerstedde, J. M. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science 295, 1301–1306 (2002).
Saunders, H. L. & Magor, B. G. Cloning and expression of the AID gene in the channel catfish. Dev. Comp. Immunol. 28, 657–663 (2004).
Nakamuta, M. et al. Complete phenotypic characterization of Apobec-1 knockout mice with a wild-type genetic background and a human apolipoprotein B transgenic background, and restoration of apolipoprotein B mRNA editing by somatic gene transfer of Apobec-1. J. Biol. Chem. 271, 25981–25988 (1996).
Muramatsu, M. et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102, 553–563 (2000).
Revy, P. et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the hyper-IgM syndrome (HIGM2). Cell 102, 565–575 (2000). References 77 and 78 show that AID is required for immunoglobulin-gene class-switch recombination and somatic hypermutation.
Muramatsu, M. et al. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 274, 18470–18476 (1999). A landmark paper that identifies AID as a B-cell-specific factor, which could be upregulated in cells induced to undergo class-switch recombination.
Yu, K., Huang, F. T. & Lieber, M. R. DNA substrate length and surrounding sequence affect the activation-induced deaminase activity at cytidine. J. Biol. Chem. 279, 6496–6500 (2004).
Sohail, A. et al. Human activation-induced cytidine deaminase causes transcription-dependent, strand-biased C to U deaminations. Nucleic Acids Res. 31, 2990–2994 (2003).
Dickerson, S. K. et al. AID mediates hypermutation by deaminating single stranded DNA. J. Exp. Med. 197, 1291–1296 (2003).
Chaudhuri, J. et al. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature 422, 726–730 (2003).
Bransteitter, R. et al. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl Acad. Sci. USA 100, 4102–4107 (2003).
Lindahl, T. & Wood, R. D. Quality control by DNA repair. Science 286, 1897–1905 (1999).
Imai, K. et al. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nature Immunol. 4, 1023–1028 (2003).
Rada, C. et al. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 12, 1748–1755 (2002).
Di Noia, J. & Neuberger, M. S. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature 419, 43–48 (2002).
Di Noia, J. M. & Neuberger, M. S. Immunoglobulin gene conversion in chicken DT40 cells largely proceeds through an abasic site intermediate generated by excision of the uracil produced by AID-mediated deoxycytidine deamination. Eur. J. Immunol. 34, 504–508 (2004).
Hochegger, H., Sonoda, E. & Takeda, S. Post-replication repair in DT40 cells: translesion polymerases versus recombinases. Bioessays 26, 151–158 (2004).
Bertocci, B. et al. DNA polymerases μ and λ are dispensable for Ig gene hypermutation. J. Immunol. 168, 3702–3706 (2002).
Petersen, S. et al. AID is required to initiate Nbs1/γ-H2AX focus formation and mutations at sites of class switching. Nature 414, 660–665 (2001).
Lahdesmaki, A. et al. Delineation of the role of the Mre11 complex in class switch recombination. J. Biol. Chem. 279, 16479–16487 (2004).
Begum, N. A. et al. Uracil DNA glycosylase activity is dispensable for immunoglobulin class switch. Science 305, 1160–1163 (2004).
Galagan, J. E. & Selker, E. U. RIP: the evolutionary cost of genome defense. Trends Genet. 20, 417–423 (2004).
Chester, A. et al. RNA editing: cytidine to uridine conversion in apolipoprotein B mRNA. Biochim. Biophys. Acta 1494, 1–13 (2000).
Anant, S. & Davidson, N. O. Molecular mechanisms of apolipoprotein B mRNA editing. Curr. Opin. Lipidol. 12, 159–165 (2001).
Petersen-Mahrt, S. K. & Neuberger, M. S. In vitro deamination of cytosine to uracil in single-stranded DNA by apolipoprotein B editing complex catalytic subunit 1 (APOBEC1). J. Biol. Chem. 278, 19583–19586 (2003).
Yamanaka, S. et al. Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc. Natl Acad. Sci. USA 92, 8483–8487 (1995).
Yamanaka, S. et al. Hyperediting of multiple cytidines of apolipoprotein B mRNA by APOBEC-1 requires auxiliary protein(s) but not a mooring sequence motif. J. Biol. Chem. 271, 11506–11510 (1996).
Kunkel, T. A. & Diaz, M. Enzymatic cytosine deamination: friend and foe. Mol. Cell 10, 962–963 (2002).
Okazaki, I. M. et al. Constitutive expression of AID leads to tumorigenesis. J. Exp. Med. 197, 1173–1181 (2003).
Loeb, L. A. et al. Lethal mutagenesis of HIV with mutagenic nucleoside analogs. Proc. Natl Acad. Sci. USA 96, 1492–1497 (1999).
Harris, R. S. et al. DNA deamination: not just a trigger for antibody diversification but also a mechanism for defense against retroviruses. Nature Immunol. 4, 641–643 (2003).
Nei, M. & Glazko, G. V. The Wilhelmine E. Key 2001 Invitational Lecture. Estimation of divergence times for a few mammalian and several primate species. J. Hered. 93, 157–164 (2002).
Madsen, P. et al. Psoriasis upregulated phorbolin-1 shares structural but not functional similarity to the mRNA-editing protein Apobec-1. J. Invest. Dermatol. 113, 162–169 (1999).
Acknowledgements
We are grateful to our laboratory and neighbouring colleagues for helpful comments, especially E. Hendrickson, D. Livingston, P. Magee and L. Mansky. We also thank T. Floss and the reviewers for helpful comments. R.S.H. is supported by a Burroughs–Wellcome Fund Hitchings Elion Fellowship (United States), the Searle Scholars Program (United States) and a new laboratory start-up award from the University of Minnesota (United States).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- TARGET CELL
-
Any cell lacking the virus under consideration that subsequently will be infected.
- APOBEC
-
(Apolipoprotein B (APOB) mRNA-editing, catalytic polypeptide). Although originally used to describe APOBEC1, which edits C6666 in APOB mRNA, this acronym has also become the accepted prefix for naming related vertebrate proteins that have the hallmark cytosine-deaminase motif His-X-Glu–X23–28-Pro-Cys-X2–4-Cys.
- DEAMINATION
-
Removal of an amine group from a pyrimidine or purine nucleic-acid base. Deamination of cytosine and adenosine yields uracil and inosine, respectively.
- PRODUCER CELL
-
Any cell used to propagate viruses.
- RETROVIRUS
-
A class of enveloped RNA virus that is distinguished by the requirement for reverse transcription of the RNA genome by a viral reverse-transcriptase enzyme to form a DNA intermediate that is then stably integrated into host chromosomal DNA. Lentiviruses such as HIV-1 are a subset of the retrovirus family that are further distinguished by numerous accessory proteins (for example, virion infectivity factor, Vif).
- MUTATIONS
-
Heritable changes in an organism's nucleic acid.
- ACTIVATION-INDUCED DEAMINASE
-
(AID). A cytosine deaminase that catalyses a pivotal step in immunoglobulin gene-diversification reactions.
- SOMATIC HYPERMUTATION
-
High-frequency base-substitution mutations found in B-cell immunoglobulin-gene variable regions.
- GENE CONVERSION
-
A non-reciprocal recombination event between homologous or partially homologous DNA sequences, leading to the templated replacement of one sequence with the other.
- CLASS-SWITCH RECOMBINATION
-
(Class or isotype switching). A region-specific recombination process, which occurs in antigen-activated B cells. This occurs between 'switch region' DNA sequences and results in a change from the IgM class to IgG, IgA or IgE. This imparts flexibility to the humoral immune response and allows it to exploit the different capacities of the immunoglobulins to activate the appropriate downstream effector mechanisms.
- RETROVIRAL HYPERMUTATION
-
The high-frequency accumulation of mutations in a retroviral genome. They are distinguished from reverse-transcriptase-dependent mutations in that they are predominantly plus-strand G to A substitutions (C to T in the minus strand).
- HYPERMUTATION
-
Levels of mutation significantly above spontaneous levels for a given system. Hypermutations are often characterized by specific local sequence preferences (biases).
- MUTATOR
-
A protein that actively promotes mutation. This is not to be confused with DNA-repair proteins, which, with the exception of the Y-family of error-prone DNA polymerases, work to discourage mutation.
- UBIQUITIN LIGASE
-
An enzyme (E2 or E3) that catalyses the transfer of ubiquitin — an ∼80-residue protein that is highly conserved among all living organisms — to a specific target protein, thereby modifying its function or marking it for degradation by the proteasome.
Rights and permissions
About this article
Cite this article
Harris, R., Liddament, M. Retroviral restriction by APOBEC proteins. Nat Rev Immunol 4, 868–877 (2004). https://doi.org/10.1038/nri1489
Issue Date:
DOI: https://doi.org/10.1038/nri1489
This article is cited by
-
Mesoscale DNA features impact APOBEC3A and APOBEC3B deaminase activity and shape tumor mutational landscapes
Nature Communications (2024)
-
The role of APOBEC3B in lung tumor evolution and targeted cancer therapy resistance
Nature Genetics (2024)
-
Epigenetic regulation in hematopoiesis and its implications in the targeted therapy of hematologic malignancies
Signal Transduction and Targeted Therapy (2023)
-
Regulatory variants of APOBEC3 genes potentially associate with COVID-19 severity in populations with African ancestry
Scientific Reports (2023)
-
APOBEC3B drives PKR-mediated translation shutdown and protects stress granules in response to viral infection
Nature Communications (2023)