Key Points
-
Analyses of cancer genome sequences and structures provide insights for understanding cancer biology, diagnosis and therapy.
-
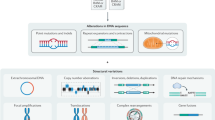
The application of second-generation DNA sequencing technologies (also known as next-generation sequencing) is allowing substantial advances in cancer genomics. In recent years, it has become feasible to sequence the expressed genes ('transcriptomes'), known exons ('exomes'), and complete genomes of cancer samples.
-
There are particular challenges for the detection and diagnosis of cancer genome alterations. For example, some cancer genome alterations are prevalent at low frequency in clinical samples, often owing to substantial admixture with non-malignant cells.
-
The large quantity of data from second-generation sequencing provides statistical and computational challenges.
-
An impetus for studies of somatic genome alterations is the potential for therapies targeted against the products of these alterations.
Abstract
Cancers are caused by the accumulation of genomic alterations. Therefore, analyses of cancer genome sequences and structures provide insights for understanding cancer biology, diagnosis and therapy. The application of second-generation DNA sequencing technologies (also known as next-generation sequencing) — through whole-genome, whole-exome and whole-transcriptome approaches — is allowing substantial advances in cancer genomics. These methods are facilitating an increase in the efficiency and resolution of detection of each of the principal types of somatic cancer genome alterations, including nucleotide substitutions, small insertions and deletions, copy number alterations, chromosomal rearrangements and microbial infections. This Review focuses on the methodological considerations for characterizing somatic genome alterations in cancer and the future prospects for these approaches.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Bentley, D. R. et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456, 53–59 (2008).
Drmanac, R. et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 327, 78–81 (2010).
Margulies, M. et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380 (2005).
Shendure, J. et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science 309, 1728–1732 (2005).
Wheeler, D. A. et al. The complete genome of an individual by massively parallel DNA sequencing. Nature 452, 872–876 (2008).
Maher, C. A. et al. Transcriptome sequencing to detect gene fusions in cancer. Nature 458, 97–101 (2009). This paper demonstrates the power of second-generation transcriptome sequencing to identify rearrrangements in coding genes.
Maher, C. A. et al. Chimeric transcript discovery by paired-end transcriptome sequencing. Proc. Natl Acad. Sci. USA 106, 12353–12358 (2009).
Ng, S. B. et al. Exome sequencing identifies the cause of a Mendelian disorder. Nature Genet. 42, 30–35 (2010).
Ng, S. B. et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461, 272–276 (2009).
Ley, T. J. et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 456, 66–72 (2008). This is the first publication describing whole-genome sequencing of a human cancer.
Mardis, E. R. et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 361, 1058–1066 (2009).
Pleasance, E. D. et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463, 191–196 (2010).
Pleasance, E. D. et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 463, 184–190 (2010).
Lee, W. et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature 465, 473–477 (2010).
Shah, S. P. et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature 461, 809–813 (2009).
Weir, B., Zhao, X. & Meyerson, M. Somatic alterations in the human cancer genome. Cancer Cell 6, 433–438 (2004).
Mitsudomi, T. et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised Phase 3 trial. Lancet Oncol. 11, 121–128 (2009).
Mok, T. S. et al. Gefitinib or carboplatin–paclitaxel in pulmonary adenocarcinoma. N. Engl. J. Med. 361, 947–957 (2009).
Rosell, R. et al. Screening for epidermal growth factor receptor mutations in lung cancer. N. Engl. J. Med. 361, 958–967 (2009).
Stratton, M. R., Campbell, P. J. & Futreal, P. A. The cancer genome. Nature 458, 719–724 (2009).
Thomas, R. K. et al. Sensitive mutation detection in heterogeneous cancer specimens by massively parallel picoliter reactor sequencing. Nature Med. 12, 852–855 (2006).
Campbell, P. J. et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nature Genet. 40, 722–729 (2008).
Feng, H., Shuda, M., Chang, Y. & Moore, P. S. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319, 1096–1100 (2008).
MacConaill, L. & Meyerson, M. Adding pathogens by genomic subtraction. Nature Genet. 40, 380–382 (2008).
Weber, G., Shendure, J., Tanenbaum, D. M., Church, G. M. & Meyerson, M. Identification of foreign gene sequences by transcript filtering against the human genome. Nature Genet. 30, 141–142 (2002).
Chiang, D. Y. et al. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nature Methods 6, 99–103 (2009).
Getz, G. et al. Comment on “The consensus coding sequences of human breast and colorectal cancers”. Science 317, 1500 (2007).
The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 (2008). This is the first paper from The Cancer Genome Atlas, which demonstrates the power of integrative analysis of multiple platforms for genomic analysis on a large series of cancer samples.
Pinard, R. et al. Assessment of whole genome amplification-induced bias through high-throughput, massively parallel whole genome sequencing. BMC Genomics 7, 216 (2006).
Gilbert, M. T. et al. The isolation of nucleic acids from fixed, paraffin-embedded tissues-which methods are useful when? PLoS ONE 2, e537 (2007).
Wood, H. M. et al. Using next-generation sequencing for high resolution multiplex analysis of copy number variation from nanogram quantities of DNA from formalin-fixed paraffin-embedded specimens. Nucleic Acids Res. 38, e151 (2010).
Gallegos Ruiz, M. I. et al. EGFR and K-ras mutation analysis in non-small cell lung cancer: comparison of paraffin embedded versus frozen specimens. Cell Oncol. 29, 257–264 (2007).
Marchetti, A., Felicioni, L. & Buttitta, F. Assessing EGFR mutations. N. Engl. J. Med. 354, 526–528 (2006).
Navin, N. et al. Inferring tumor progression from genomic heterogeneity. Genome Res. 20, 68–80 (2010).
Ding, L. et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464, 999–1005 (2010). The first publication of the comprehensive sequencing of primary and metastatic tumour material from an individual.
Shendure, J. & Ji, H. Next-generation DNA sequencing. Nature Biotech. 26, 1135–1145 (2008).
Pettersson, E., Lundeberg, J. & Ahmadian, A. Generations of sequencing technologies. Genomics 93, 105–111 (2009).
Hoffman, B. G. & Jones, S. J. Genome-wide identification of DNA–protein interactions using chromatin immunoprecipitation coupled with flow cell sequencing. J. Endocrinol. 201, 1–13 (2009).
Stephens, P. J. et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 462, 1005–1010 (2009). This is the largest collection of samples for a single cancer type to be subject to whole-genome rearrangement analysis and documents the large sample-to-sample variability in frequency of events.
Rowley, J. D. Chromosome translocations: dangerous liaisons revisited. Nature Rev. Cancer 1, 245–250 (2001).
Meyerson, M. Cancer: broken genes in solid tumours. Nature 448, 545–546 (2007).
Tomlins, S. A. et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310, 644–648 (2005).
Soda, M. et al. Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature 448, 561–566 (2007).
Beck, C. R. et al. LINE-1 retrotransposition activity in human genomes. Cell 141, 1159–1170 (2010).
Huang, C. R. et al. Mobile interspersed repeats are major structural variants in the human genome. Cell 141, 1171–1182 (2010).
Albert, T. J. et al. Direct selection of human genomic loci by microarray hybridization. Nature Methods 4, 903–905 (2007).
Gnirke, A. et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nature Biotech. 27, 182–189 (2009).
Hodges, E. et al. Genome-wide in situ exon capture for selective resequencing. Nature Genet. 39, 1522–1527 (2007).
Turner, E. H., Lee, C., Ng, S. B., Nickerson, D. A. & Shendure, J. Massively parallel exon capture and library-free resequencing across 16 genomes. Nature Methods 6, 315–316 (2009).
Levin, J. Z. et al. Targeted next-generation sequencing of a cancer transcriptome enhances detection of sequence variants and novel fusion transcripts. Genome Biol. 10, R115 (2009).
Davies, H. et al. Mutations of the BRAF gene in human cancer. Nature 417, 949–954 (2002).
Paez, J. G. et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304, 1497–1500 (2004).
Lynch, T. J. et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 350, 2129–2139 (2004).
Pao, W. et al. EGF receptor gene mutations are common in lung cancers from 'never smokers' and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl Acad. Sci. USA 101, 13306–13311 (2004). References 52–54 were the first publications to link therapeutic outcome in lung cancer to specific somatically acquired point mutations, and they suggest the value of systematic sequencing of kinase gene families.
Stephens, P. et al. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature 431, 525–526 (2004).
Baxter, E. J. et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet 365, 1054–1061 (2005).
James, C. et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 434, 1144–1148 (2005).
Kralovics, R. et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N. Engl. J. Med. 352, 1779–1790 (2005).
Levine, R. L. et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell 7, 387–397 (2005).
Zhao, R. et al. Identification of an acquired JAK2 mutation in polycythemia vera. J. Biol. Chem. 280, 22788–22792 (2005).
Dutt, A. et al. Drug-sensitive FGFR2 mutations in endometrial carcinoma. Proc. Natl Acad. Sci. USA 105, 8713–8717 (2008).
Pollock, P. M. et al. Frequent activating FGFR2 mutations in endometrial carcinomas parallel germline mutations associated with craniosynostosis and skeletal dysplasia syndromes. Oncogene 26, 7158–7162 (2007).
Chen, Y. et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature 455, 971–974 (2008).
George, R. E. et al. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature 455, 975–978 (2008).
Janoueix-Lerosey, I. et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature 455, 967–970 (2008).
Mosse, Y. P. et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature 455, 930–935 (2008).
Samuels, Y. et al. High frequency of mutations of the PIK3CA gene in human cancers. Science 304, 554 (2004).
Jones, S. et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321, 1801–1806 (2008).
Parsons, D. W. et al. An integrated genomic analysis of human glioblastoma multiforme. Science 321, 1807–1812 (2008).
Sjoblom, T. et al. The consensus coding sequences of human breast and colorectal cancers. Science 314, 268–274 (2006). This paper described the first example of whole-exome sequencing of human cancers.
Wood, L. D. et al. The genomic landscapes of human breast and colorectal cancers. Science 318, 1108–1113 (2007).
Jones, S. et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science 324, 217 (2009).
Bainbridge, M. N. et al. Whole exome capture in solution with 3 Gbp of data. Genome Biol. 11, R62 (2010).
Thomas, R. K. et al. High-throughput oncogene mutation profiling in human cancer. Nature Genet. 39, 347–351 (2007).
Berger, M. F. et al. Integrative analysis of the melanoma transcriptome. Genome Res. 20, 413–427 (2010).
Palanisamy, N. et al. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nature Med. 16, 793–798 (2010).
Shah, S. P. et al. Mutation of FOXL2 in granulosa-cell tumors of the ovary. N. Engl. J. Med. 360, 2719–2729 (2009).
Morrissy, A. S. et al. Next-generation tag sequencing for cancer gene expression profiling. Genome Res. 19, 1825–1835 (2009).
Ding, L. et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455, 1069–1075 (2008).
Goya, R. et al. SNVMix: predicting single nucleotide variants from next-generation sequencing of tumors. Bioinformatics 26, 730–736 (2010).
Ng, P. C. & Henikoff, S. Predicting deleterious amino acid substitutions. Genome Res. 11, 863–874 (2001).
Kaminker, J. S., Zhang, Y., Watanabe, C. & Zhang, Z. CanPredict: a computational tool for predicting cancer-associated missense mutations. Nucleic Acids Res. 35, W595–W598 (2007).
Adzhubei, I. A. et al. A method and server for predicting damaging missense mutations. Nature Methods 7, 248–249 (2010).
Ramensky, V., Bork, P. & Sunyaev, S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 30, 3894–3900 (2002).
Carter, H. et al. Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations. Cancer Res. 69, 6660–6667 (2009).
Hahn, W. C. & Weinberg, R. A. Rules for making human tumor cells. N. Engl. J. Med. 347, 1593–1603 (2002).
Beroukhim, R. et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc. Natl Acad. Sci. USA 104, 20007–20012 (2007).
Beroukhim, R. et al. The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010). This paper is an analysis of somatic copy number changes across 26 different human cancer types and points to regions commonly altered at significant levels across cancer types.
Bignell, G. R. et al. Signatures of mutation and selection in the cancer genome. Nature 463, 893–898 (2010).
Bignell, G. R. et al. High-resolution analysis of DNA copy number using oligonucleotide microarrays. Genome Res. 14, 287–295 (2004).
Mullighan, C. G. et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 446, 758–764 (2007).
Weir, B. A. et al. Characterizing the cancer genome in lung adenocarcinoma. Nature 450, 893–898 (2007).
Zhao, X. et al. An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res. 64, 3060–3071 (2004).
Zhao, X. et al. Homozygous deletions and chromosome amplifications in human lung carcinomas revealed by single nucleotide polymorphism array analysis. Cancer Res. 65, 5561–5570 (2005).
Tengs, T. et al. Genomic representations using concatenates of type IIB restriction endonuclease digestion fragments. Nucleic Acids Res. 32, e121 (2004).
Wang, T. L. et al. Digital karyotyping. Proc. Natl Acad. Sci. USA 99, 16156–16161 (2002).
Velculescu, V. E., Zhang, L., Vogelstein, B. & Kinzler, K. W. Serial analysis of gene expression. Science 270, 484–487 (1995).
Chen, K. et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nature Methods 6, 677–681 (2009).
Leary, R. J. et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci. Transl. Med. 2, 20ra14 (2010).
Dalla-Favera, R. et al. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc. Natl Acad. Sci. USA 79, 7824–7827 (1982).
Durst, M., Gissmann, L., Ikenberg, H. & zur Hausen, H. A papillomavirus DNA from a cervical carcinoma and its prevalence in cancer biopsy samples from different geographic regions. Proc. Natl Acad. Sci. USA 80, 3812–3815 (1983).
Feng, H. et al. Human transcriptome subtraction by using short sequence tags to search for tumor viruses in conjunctival carcinoma. J. Virol. 81, 11332–11340 (2007).
Xu, Y. et al. Pathogen discovery from human tissue by sequence-based computational subtraction. Genomics 81, 329–335 (2003).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Koboldt, D. C. et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25, 2283–2285 (2009).
Ye, K., Schulz, M. H., Long, Q., Apweiler, R. & Ning, Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25, 2865–2871 (2009).
McKenna, A. H. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 19 Jul 2010 (doi:10.1101/gr.107524.110).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589–595 (2010).
Ning, Z., Cox, A. J. & Mullikin, J. C. SSAHA: a fast search method for large DNA databases. Genome Res. 11, 1725–1729 (2001).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Li, R. et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967 (2009).
Rumble, S. M. et al. SHRiMP: accurate mapping of short color-space reads. PLoS Comput. Biol. 5, e1000386 (2009).
Homer, N., Merriman, B. & Nelson, S. F. BFAST: an alignment tool for large scale genome resequencing. PLoS ONE 4, e7767 (2009).
LaFramboise, T. et al. Allele-specific amplification in cancer revealed by SNP array analysis. PLoS Comput. Biol. 1, e65 (2005).
Maheswaran, S. et al. Detection of mutations in EGFR in circulating lung-cancer cells. N. Engl. J. Med. 359, 366–377 (2008).
Bentley, D. R. et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456, 53–59 (2008).
Venkatraman, E. S. & Olshen, A. B. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics 23, 657–663 (2007).
Reva, B., Antipin, Y. & Sander, C. Determinants of protein function revealed by combinatorial entropy optimization. Genome Biol. 8, R232 (2007).
Krzywinski, M. et al. Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009).
Acknowledgements
We thank M. Lawrence and G. Saksena for careful review of the manuscript. We acknowledge support from The Cancer Genome Atlas programme of the National Cancer Institute, U24CA143867 and U24CA143845, and from the National Human Genome Research Institute, U54HG003067.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Matthew Meyerson receives research support from Genentech, is a consultant to and receives research support from Novartis, and is a founding advisor of and a consultant to Foundation Medicine.
Glossary
- Second-generation sequencing
-
Used in this Review to refer to sequencing methods that have emerged since 2005 that parallelize the sequencing process and produce millions of typically short sequence reads (50–400 bases) from amplified DNA clones. It is also often known as next-generation sequencing.
- First-generation sequencing
-
(also known as Sanger sequencing or capillary sequencing). The standard sequencing methodology used to sequence the reference human (and other model organism) genomes. It uses radioactively or fluorescently labelled dideoxynucleotide triphosphates (ddNTPs) as DNA chain terminators. Various detection methods allow read-out of sequence according to the incorporation of each specific terminator (ddATP, ddCTP, ddGTP or ddTTP).
- Whole-genome amplification
-
Various molecular techniques (including multiple displacement amplification, rolling circle amplification or degenerate oligonucleotide primed PCR) in which very small amounts (nanograms) of a genomic DNA sample can be multiplied in a largely unbiased fashion to produce suitable quantities for genomic analysis (micrograms).
- Moore's law
-
The observation made in 1965 by Gordon Moore that the number of transistors per square inch on integrated circuits had doubled every other year since the integrated circuit was invented.
- Chromatin immunoprecipitation
-
A technique used to identify the location of DNA-binding proteins and epigenetic marks in the genome. Genomic sequences containing the protein of interest are enriched by binding soluble DNA chromatin extracts (complexes of DNA and protein) to an antibody that recognizes the protein or modification.
- Over-sampling
-
Reading the same stretch of DNA sequence many times to gain a confident sequence read-out.
- Shotgun sequencing
-
Sequencing randomly derived fragments of the whole genome. The order and orientation of the sequences are determined by mapping individual reads back to a reference or through assembly of overlapping sequences into larger contigs of sequence.
- Jumping library
-
A method of library construction in which the genome is divided into large fragments using a rare cutter enzyme. Fragments are circularized and DNA sequences are read from the ends of the fragment, without reading the intervening sequence.
- Transformation assay
-
The measurement of cell phenotypes to assess oncogenic changes.
- Digital karyotyping
-
A method to quantify DNA copy number. Short sequence-derived tags that cover the genome are used to read-out relative copy number.
- Directed sequencing
-
Sequencing only subsets of the genome, for example, particular genes or regions of interest.
- Free serum DNA
-
DNA that is cell-free and is circulating in the bloodstream. It typically refers to tumour DNA that can be isolated in the blood.
Rights and permissions
About this article
Cite this article
Meyerson, M., Gabriel, S. & Getz, G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet 11, 685–696 (2010). https://doi.org/10.1038/nrg2841
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg2841
This article is cited by
-
Comprehensive genomic profiling for oncological advancements by precision medicine
Medical Oncology (2023)
-
Whole-exome sequencing identified mutational profiles of urothelial carcinoma post kidney transplantation
Journal of Translational Medicine (2022)
-
Duplex sequencing identifies genomic features that determine susceptibility to benzo(a)pyrene-induced in vivo mutations
BMC Genomics (2022)
-
Application of second-generation sequencing in congenital pulmonary airway malformations
Scientific Reports (2022)