Key Points
-
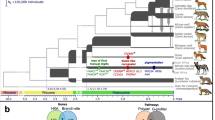
The dog is descended from a single species, the grey wolf, but from an ancient lineage that is now extinct and is the only large carnivore ever to be domesticated.
-
Dogs were domesticated by hunter-gatherers over 15,000 years ago, whereas all other domesticated species were domesticated by agrarian societies.
-
The demographic history of the dog is complex, with multiple population bottlenecks associated with domestication and breed formation as well as admixture among dog populations and with wolves.
-
DNA analysis from modern dogs can be used to recapitulate the historic migration of human populations, providing previously unknown information about how various regions were settled.
-
Whole-genome sequencing of modern dogs has generated a large catalogue that captures much of the variation that exists in modern dogs.
-
There are nearly 400 breeds of dog worldwide that display an extraordinary amount of phenotypic diversity in terms of morphology, behaviour, and disease susceptibility. All domesticated dogs are members of the same species, termed Canis lupus familiaris or, alternatively, Canis familiaris.
-
Selection for behaviour and appearance in modern breeds has resulted in an increased level of disease susceptibility, perhaps as deleterious alleles 'hitch-hike' with those that breeders and fanciers select for.
-
Cancer is an extremely common disease in dogs, and findings from breed-specific studies reveal information that is relevant for basic cancer biology.
Abstract
The domestic dog represents one of the most dramatic long-term evolutionary experiments undertaken by humans. From a large wolf-like progenitor, unparalleled diversity in phenotype and behaviour has developed in dogs, providing a model for understanding the developmental and genomic mechanisms of diversification. We discuss pattern and process in domestication, beginning with general findings about early domestication and problems in documenting selection at the genomic level. Furthermore, we summarize genotype–phenotype studies based first on single nucleotide polymorphism (SNP) genotyping and then with whole-genome data and show how an understanding of evolution informs topics as different as human history, adaptive and deleterious variation, morphological development, ageing, cancer and behaviour.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Wayne, R. K. Cranial morphology of domestic and wild canids the influence of development on morphological change. Evolution 40, 243–261 (1986).
Wayne, R. K. Limb morphology of domestic and wild canids: the influence of development on morphologic change. J. Morphol. 187, 301–319 (1986).
Drake, A. G. & Klingenberg, C. P. Large-scale diversification of skull shape in domestic dogs: disparity and modularity. Am. Nat. 175, 289–301 (2010).
Parker, H. G., Shearin, A. L. & Ostrander, E. A. Man's best friend becomes biology's best in show: genome analyses in the domestic dog. Annu. Rev. Genet. 44, 309–336 (2010).
vonHoldt, B. M. et al. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature 464, 898–902 (2010). The authors present a genetic similarity tree of dog breeds that suggests breed grouping by form and function and implies that cross breeding between these breed groups transfers key genetic mutations that are responsible for discrete phenotypic traits. They identify genes that may have been under selection in the origin of dogs.
Thalmann, O. et al. Complete mitochondrial genomes of ancient canids suggest a European origin of domestic dogs. Science 342, 871–874 (2013). For the first time, ancient DNA of dogs and wolves was used to illuminate the timing and geographic origin of dogs, and it suggests that dogs originated from an extinct wolf population that existed in Europe about 27,000 years ago.
Shannon, L. M. et al. Genetic structure in village dogs reveals a Central Asian domestication origin. Proc. Natl Acad. Sci. USA 112, 13639–13644 (2015).
Skoglund, P., Ersmark, E., Palkopoulou, E. & Dalen, L. Ancient wolf genome reveals an early divergence of domestic dog ancestors and admixture into high-latitude breeds. Curr. Biol. 25, 1515–1519 (2015).
Wang, G. D. et al. Out of southern East Asia: the natural history of domestic dogs across the world. Cell Res. 26, 21–33 (2016).
Fan, Z. et al. Worldwide patterns of genomic variation and admixture in gray wolves. Genome Res. 26, 163–173 (2016).
Frantz, L. A. et al. Genomic and archaeological evidence suggest a dual origin of domestic dogs. Science 352, 1228–1231 (2016).
Ash, E. C. Dogs: Their History and Development (E. Benn Limited, 1927).
American Kennel Club. The Complete Dog Book 20th edn (Ballantine Books, 2006).
Parker, H. G. et al. Genetic structure of the purebred domestic dog. Science 304, 1160–1164 (2004).
Lindblad-Toh, K. et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438, 803–819 (2005). This paper announced the first whole-genome sequence of a dog, the Boxer, together with a description of unique elements within the canine genome and predictions of ancestral sample size and SNP density needed for whole-genome association studies.
Boyko, A. R. et al. Complex population structure in African village dogs and its implications for inferring dog domestication history. Proc. Natl Acad. Sci. USA 106, 13903–13908 (2009).
Freedman, A. H. et al. Genome sequencing highlights the dynamic early history of dogs. PLoS Genet. 10, e1004016 (2014). In this paper, the authors compare whole-genome sequences from several canine lineages, demonstrating that a severe bottleneck occurred in wolves soon after their divergence from dogs, implying that the pool of diversity that gave rise to dogs was substantially larger than that represented by modern wolf populations and narrowing the time frame for initial dog domestication to 11–16 thousand years ago.
Freedman, A. H. et al. Demographically-based evaluation of genomic regions under selection in domestic dogs. PLoS Genet. 12, e1005851 (2016). Explicit demographic models based on genetic data were used to develop a null distribution and better reduce the false positives for selective sweeps. The work shows selection on genes related to neurological, metabolic and phenotypic traits, but it does not confirm many of the genes proposed to be under selection in previous studies.
Marsden, C. D. et al. Bottlenecks and selective sweeps during domestication have increased deleterious genetic variation in dogs. Proc. Natl Acad. Sci. USA 113, 152–157 (2016).
Dreger, D. L. et al. Whole-genome sequence, SNP chips and pedigree structure: building demographic profiles in domestic dog breeds to optimize genetic-trait mapping. Dis. Model. Mech. 9, 1445–1460 (2016).
Sutter, N. B. et al. Extensive and breed-specific linkage disequilibrium in Canis familiaris. Genome Res. 14, 2388–2396 (2004).
Boyko, A. et al. A simple genetic architecture underlies morphological variation in dogs. PLoS Biol. 8, e1000451 (2010).
Akey, J. M. et al. Tracking footprints of artificial selection in the dog genome. Proc. Natl Acad. Sci. USA 107, 1160–1165 (2010).
Vaysse, A. et al. Identification of genomic regions associated with phenotypic variation between dog breeds using selection mapping. PLoS Genet. 7, e1002316 (2011).
Hoopes, B. C., Rimbault, M., Liebers, D., Ostrander, E. A. & Sutter, N. B. The insulin-like growth factor 1 receptor (IGF1R) contributes to reduced size in dogs. Mamm. Genome 23, 780–790 (2012).
Axelsson, E. et al. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature 495, 360–364 (2013). These investigators used whole-genome resequencing of wolves and dogs to identify genes under selection during domestication, showing for the first time that amplifications of the amylase gene in early dogs provided a mechanism to survive on a starch-based diet.
Haywood, S. et al. Copper toxicosis in non-COMMD1 Bedlington terriers is associated with metal transport gene ABCA12. J. Trace Elem. Med. Biol. 35, 83–89 (2016).
Cagan, A. & Blass, T. Identification of genomic variants putatively targeted by selection during dog domestication. BMC Evol. Biol. 16, 10 (2016).
Decker, B. et al. Comparison against 186 canid whole-genome sequences reveals survival strategies of an ancient clonally transmissible canine tumor. Genome Res. 25, 1646–1655 (2015). This work describes the use of the largest multibreed whole-genome sequence data set to date, for filtering potential deleterious variants from millions to thousands and subsequent reduction to cancer pathways that are responsible for transmissible tumours.
Freedman, A. H. & Wayne, R. K. Deciphering the origin of dogs: from fossils to genomes. Annu. Rev. Anim. Biosci. 5, 281–307 (2017).
Barton, N. H. The effect of hitch-hiking on neutral genealogies. Genet. Res. 72, 123–133 (1998).
Barton, N. H. Genetic hitchhiking. Phil. Trans. R. Soc. Lond. B 355, 1553–1562 (2000).
Crisci, J. L., Poh, Y. P., Mahajan, S. & Jensen, J. D. The impact of equilibrium assumptions on tests of selection. Front. Genet. 4, 235 (2013).
Santiago, E. & Caballero, A. Variation after a selective sweep in a subdivided population. Genetics 169, 475–483 (2005).
Slatkin, M. & Wiehe, T. Genetic hitch-hiking in a subdivided population. Genet. Res. 71, 155–160 (1998).
Arendt, M., Cairns, K. M., Ballard, J. W., Savolained, P. & Axelsson, E. Diet adaption in dog reflects spread of prehistoric agriculture. Heredity (Edinb.) 117, 301–306 (2016).
Parker, H. et al. Breed relationships facilitate fine-mapping studies: a 7.8-kb deletion cosegregates with Collie eye anomaly across multiple dog breeds. Genome Res. 17, 1652–1571 (2007).
Anderson, T. M. et al. Molecular and evolutionary history of melanism in North American gray wolves. Science 323, 1339–1343 (2009).
Li, Y. et al. Population variation revealed high-altitude adaptation of Tibetan mastiffs. Mol. Biol. Evol. 31, 1200–1205 (2014).
vonHoldt, B., Fan, Z., Ortega-Del Vecchyo, D. & Wayne, R. K. EPAS1 variants in high altitude Tibetan wolves were selectively introgressed into highland dogs. PeerJ 5, e3522 (2017).
Lin, L., Vad-Nielsen, J. & Luo, Y. CRISPR-mediated multiplexed genetic manipulation. Oncotarget 7, 80103–80104 (2016).
Shipman, P. The Invaders: How Humans and Their Dogs Drove Neanderthals to Extinction 3rd edn (Belknap Press, 2015).
Tonoike, A. et al. Copy number variations in the amylase gene (AMY2B) in Japanese native dog breeds. Anim. Genet. 46, 580–583 (2015).
Ollivier, M. et al. Amy2B copy number variation reveals starch diet adaptations in ancient European dogs. R. Soc. Open Sci. 3, 160449 (2016). This paper presents ancient DNA data mapping the temporal origin of the increased amylase gene copy number using DNA from ancient dogs and shows that it first appeared over 7,000 years ago, concurrent with the beginning of agriculture.
Inchley, C. E. et al. Selective sweep on human amylase genes postdates the split with Neanderthals. Sci. Rep. 6, 37198 (2016).
Reiter, T., Jagoda, E. & Capellini, T. D. Dietary variation and evolution of gene copy number among dog breeds. PLoS ONE 11, e0148899 (2016).
Botigue, L. et al. Ancient European dog genomes reveal continuity since the early Neolithic. Nat. Commun. 8, 16082 (2017).
Dreger, D. L. et al. Commonalities in development of pure breeds and population isolates revealed in the genome of the sardinian Fonni's Dog. Genetics 204, 737–755 (2016).
Cetti, F. I quadrupedi di Sardegna. (ed. Edoardo Perino) (Gia Sassari, 1885).
Tyndale, J. W. The Island of Sardinia, including pictures of the manners and customs of the Sardinians, and notes on the antiquities and modern objects of interst in the island: to which is added some account of the House of Savoy. Vol. 2 (Richard Bentley, 1849).
Fiorito, G. et al. The Italian genome reflects the history of Europe and the Mediterranean basin. Eur. J. Hum. Genet. 24, 1056–1062 (2015).
Gou, X. et al. Whole-genome sequencing of six dog breeds from continuous altitudes reveals adaptation to high-altitude hypoxia. Genome Res. 24, 1308–1315 (2014).
Zhang, J. E. et al. Polymorphisms in the prion protein gene (PRNP) in the Tibetan Mastiff. Anim. Genet. 40, 1001–1002 (2014).
Miao, B., Wang, Z. & Li, Y. Genomic analysis reveals hypoxia adaptation in the Tibetan mastiff by introgression of the grey wolf from the Tibetan Plateau. Mol. Biol. Evol. 34, 734–743 (2017).
Akashi, H., Osada, N. & Ohta, T. Weak selection and protein evolution. Genetics 192, 15–31 (2012).
Boyko, A. R. et al. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 4, e1000083 (2008).
Keightley, P. D. & Eyre-Walker, A. Joint inference of the distribution of fitness effects of deleterious mutations and population demography based on nucleotide polymorphism frequencies. Genetics 177, 2251–2261 (2007).
Ohta, T. Role of very slightly deleterious mutations in molecular evolution and polymorphism. Theor. Popul. Biol. 10, 254–275 (1976).
Eyre-Walker, A., Woolfit, M. & Phelps, T. The distribution of fitness effects of new deleterious amino acid mutations in humans. Genetics 173, 891–900 (2006).
Tennessen, J. A. et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 337, 64–69 (2012).
Nelson, M. R. et al. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science 337, 100–104 (2012).
Bellumori, T. P., Famula, T. R., Bannasch, D. L., Belanger, J. M. & Oberbauer, A. M. Prevalence of inherited disorders among mixed-breed and purebred dogs: 27,254 cases (1995–2010). J. Am. Vet. Med. Assoc. 242, 1549–1555 (2013).
Schoenebeck, J. J. & Ostrander, E. A. Insights into morphology and disease from the dog genome project. Annu. Rev. Cell Dev. Biol. 30, 535–560 (2014).
Karyadi, D. M. et al. A copy number variant at the KITLG locus likely confers risk for canine squamous cell carcinoma of the digit. PLoS Genet. 9, e1003409 (2013).
Ostrander, E. A., Davis, B. W. & Ostrander, G. K. Transmissible tumors: breaking the cancer paradigm. Trends Genet. 32, 1–15 (2016).
O'Neill, D. G. et al. Epidemiological associations between brachycephaly and upper respiratory tract disorders in dogs attending veterinary practices in England. Canine Genet. Epidemiol. 2, 10 (2015).
Olsson, M. et al. A novel unstable duplication upstream of HAS2 predisposes to a breed-defining skin phenotype and a periodic fever syndrome in Chinese Shar-Pei dogs. PLoS Genet. 7, e1001332 (2011).
Davis, B. W. & Ostrander, E. A. Domestic dogs and cancer research: a breed-based genomics approach. ILAR J. 55, 59–68 (2014).
Boyko, A. R. The domestic dog: man's best friend in the genomic era. Genome Biol. 12, 216 (2011).
Mirkena, T. et al. Genetics of adaptation in domestic farm animals: a review. Livestock Sci. 132, 1–12 (2010).
Bertolini, F. et al. Evidence of selection signatures that shape the Persian cat breed. Mamm. Genome 27, 144–155 (2016).
Gandolfi, B. et al. A splice variant in KRT71 is associated with curly coat phenotype of Selkirk Rex cats. Sci. Rep. 3, 2000 (2013).
Lyons, L. A. et al. Aristaless-Like Homeobox protein 1 (ALX1) variant associated with craniofacial structure and frontonasal dysplasia in Burmese cats. Dev. Biol. 409, 451–458 (2016).
Lipinski, M. J. et al. The ascent of cat breeds: genetic evaluations of breeds and worldwide random-bred populations. Genomics 91, 12–21 (2008).
Parker, H. G. et al. An expressed fgf4 retrogene is associated with breed-defining chondrodysplasia in domestic dogs. Science 325, 995–998 (2009).
Cadieu, E. et al. Coat variation in the domestic dog is governed by variants in three genes. Science 326, 150–153 (2009).
Baranowska Körberg, I. et al. A simple repeat polymorphism in the MITF-M promoter is a key regulator of white spotting in dogs. PLoS ONE 9, e104363 (2014).
Hayward, J. J. et al. Complex disease and phenotype mapping in the domestic dog. Nat. Commun. 7, 10460 (2016). Using the largest SNP-based data set to date, which encompassed 4,200 dogs genotyped with 180,000 SNPs, this paper summarizes selection data for dozens of morphological, behavioural and disease features.
Parker, H. G. et al. Genomic analyses reveal the influence of geographic origin, immigration and cross-breed introgression on modern dog breed development. Cell Rep. 19, 697–708 (2017).
Shearin, A. L. & Ostrander, E. A. Canine morphology: hunting for genes and tracking mutations. PLoS Biol. 8, e1000310 (2010).
Rimbault, M. et al. Derived variants at six genes explain nearly half of size reduction in dog breeds. Genome Res. 23, 1985–1995 (2013).
Plassais, J. et al. Analysis of large versus small dogs reveals three genes on the canine X chromosome associated with body weight, muscling and back fat thickness. PLoS Genet. 13, e1006661 (2017).
Sutter, N. B. et al. A single IGF1 allele is a major determinant of small size in dogs. Science 316, 112–115 (2007).
Vrieze, S. I. et al. An assessment of the individual and collective effects of variants on height using twins and a developmentally informative study design. PLoS Genet. 7, e1002413 (2011).
Lango, A. H. et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467, 832–838 (2010).
N'Diaye, A. et al. Identification, replication, and fine-mapping of loci associated with adult height in individuals of African ancestry. PLoS Genet. 7, e1002298 (2011).
Qu, B. H., Karas, M., Koval, A. & LeRoith, D. Insulin receptor substrate-4 enhances insulin-like growth factor-I-induced cell proliferation. J. Biol. Chem. 274, 31179–31184 (1999).
Melkersson, K. & Persson, B. Association between body mass index and insulin receptor substrate-4 (IRS-4) gene polymorphisms in patients with schizophrenia. Neuro Endocrinol. Lett. 32, 634–640 (2011).
Sun, Y. et al. Loss-of-function mutations in IGSF1 cause an X-linked syndrome of central hypothyroidism and testicular enlargement. Nat. Genet. 44, 1375–1381 (2012).
Joustra, S. D. et al. IGSF1 variants in boys with familial delayed puberty. Eur. J. Pediatr. 174, 687–692 (2015).
Asakura, Y. et al. Combined growth hormone and thyroid-stimulating hormone deficiency in a japanese patient with a novel frameshift mutation in IGSF1. Horm. Res. Paediatr. 84, 349–354 (2015).
Ma, J. et al. Fine mapping of fatness QTL on porcine chromosome X and analyses of three positional candidate genes. BMC Genet. 14, 46 (2013).
Cepica, S., Bartenschlager, H. & Geldermann, H. Mapping of QTL on chromosome X for fat deposition, muscling and growth traits in a wild boar x Meishan F2 family using a high-density gene map. Anim. Genet. 38, 634–638 (2007).
Schoenebeck, J. & Ostrander, E. A. The genetics of canine skull shape variation. Genetics 193, 317–325 (2013).
Bannasch, D. et al. Localization of canine brachycephaly using an across breed mapping approach. PLoS ONE 5, e9632 (2010).
Marchant, T. W. et al. Canine brachycephaly is associated with a retrotransposon-mediated missplicing of SMOC2. Curr. Biol. 27, 1573–1584.e6 (2017).
Schoenebeck, J. J. et al. Variation of BMP3 contributes to dog breed skull diversity. PLoS Genet. 8, e1002849 (2012).
Jones, P. et al. Single-nucleotide-polymorphism-based association mapping of dog stereotypes. Genetics 179, 1033–1044 (2008).
Holder, A., Mella, S., Palmer, D. B., Aspinall, R. & Catchpole, B. An age-associated decline in thymic output differs in dog breeds according to their longevity. PLoS ONE 11, e0165968 (2016).
Fick, L. J. et al. Telomere length correlates with life span of dog breeds. Cell Rep. 2, 1530–1536 (2012).
Bonnett, B. N., Egenvall, A., Hedhammar, A. & Olson, P. Mortality in over 350,000 insured Swedish dogs from 1995-2000: I. Breed-, gender-, age- and cause-specific rates. Acta Vet. Scand. 46, 105–120 (2005).
Kraus, C., Pavard, S. & Promislow, D. E. The size-life span trade-off decomposed: why large dogs die young. Am. Nat. 181, 492–505 (2013). This work puts forth and defends the hypothesis that small dogs live longer than large dogs because large dogs simply age at an accelerated pace once senescence is initiated.
Selman, C., Nussey, D. H. & Monaghan, P. Ageing: it's a dog's life. Curr. Biol. 23, R451–R453 (2013).
Dobson, J. M. Breed-predispositions to cancer in pedigree dogs. ISRN Vet. Sci. 2013, 941275 (2014).
Vail, D. M. & MacEwen, E. G. Spontaneously occurring tumors of companion animals as models for human cancer. Cancer Invest. 18, 781–792 (2000).
Adams, V. J., Evans, K. M., Sampson, J. & Wood, J. L. N. Methods and mortality results of a health survey of purebred dogs in the UK. J. Small Anim. Pract. 51, 512–524 (2010).
Merlo, D. F. et al. Cancer incidence in pet dogs: findings of the Animal Tumor Registry of Genoa. Italy. J. Vet. Intern. Med. 22, 976–984 (2008).
Khanna, C. et al. The dog as a cancer model. Nat. Biotechnol. 24, 1065–1066 (2006).
Dorn, C. R. Epidemiology of canine and feline tumors. Comp. Cont. Educ. Pract. Vet. 12, 307–312 (1976).
Cadieu, E. & Ostrander, E. A. Canine genetics offers new mechanisms for the study of human cancer. Cancer Epidemiol. Biomarker Prev. 16, 2181–2183 (2007).
Ranieri, G. et al. A model of study for human cancer: spontaneous occurring tumors in dogs. Biological features and translation for new anticancer therapies. Crit. Rev. Oncol. Hematol. 88, 187–197 (2013).
Dhawan, D. et al. Comparative gene expression analyses identify luminal and nasal subtypes of canine invasive urothelial carcinoma that mimic patterns in human invasive bladder cancer. PLoS ONE 10, e0136688 (2015).
Knapp, D. W., Dhawan, D. & Ostrander, E. “Lassie, ” “Toto, ” and fellow pet dogs: poised to lead the way for advances in cancer prevention. Am. Soc. Clin. Oncol. Educ. Book. http://dx.doi.org/10.14694/EdBook_AM.2015.35.e667 (2015).
Ostrander, E. A. Franklin, H. Epstein Lecture. Both ends of the leash — the human links to good dogs with bad genes. N. Engl. J. Med. 367, 636–646 (2012).
Rowell, J. L., McCarthy, D. O. & Alvarez, C. E. Dog models of naturally occurring cancer. Trends Mol. Med. 17, 380–388 (2011).
Jónasdóttir, T. J. et al. Genetic mapping of a naturally occurring hereditary renal cancer syndrome in dogs. Proc. Natl Acad. Sci. USA 97, 4132–4137 (2000).
Shearin, A. et al. The MTAP-CDKN2A locus confers susceptibility to a naturally occurring canine cancer. Cancer Epidmiol Biomarkers Prev. 21, 1019–1027 (2012).
Thomas, R., Smith, K. C., Ostrander, E. A., Galibert, F. & Breen, M. Chromosome aberrations in canine multicentric lymphomas detected with comparative genomic hybridisation and a panel of single locus probes. Br. J. Cancer 89, 1530–1537 (2003).
Phillips, J. C., Lembcke, L. & Chamberlin, T. A novel locus for canine osteosarcoma (OSA1) maps to CFA34, the canine orthologue of human 3q26. Genomics 96, 220–227 (2010).
Affolter, V. K. & Moore, P. F. Localized and disseminated histiocytic sarcoma of dendritic cell origin in dogs. Vet. Pathol. 39, 74–83 (2002).
Moore, P. F., Affolter, V. K. & Vernau, W. Canine hemophagocytic histiocytic sarcoma: a proliferative disorder of CD11d+ macrophages. Vet. Pathol. 43, 632–645 (2006).
Fulmer, A. K. & Mauldin, G. E. Canine histiocytic neoplasia: an overview. Can. Vet. J. 48, 1041–1050 (2007).
Dobson, J., Hoaher, T., McKinley, T. J. & Wood, J. L. Mortality in a cohort of flat-coated retrievers in the UK. Vet. Comp. Oncol. 7, 115–121 (2009).
Bonnett, B. N., Egenvall, A., Olson, P. & Hedhammar, A. Mortality in insured Swedish dogs: rates and causes of death in various breeds. Vet. Rec. 141, S40–S44 (1997).
Mueller, F., Fuchs, B. & Kaser-Hotz, B. Comparative biology of human and canine osteosarcoma. Anticancer Res. 27, 155–164 (2007).
Karlsson, E. et al. Genome-wide analysis implicate 33 loci in heritable dog osteosarcoma, including regulatory variants near CDKN2A/B. Genome Biol. 14, R132 (2013).
Gardner, H. L., Fenger, J. M., & London, C. A. Dogs as a model for cancer. Annu. Rev. Anim. Biosci. 4, 199–222 (2016).
Decker, B. et al. Homologous mutation to human BRAF V600E is common in naturally occurring canine bladder cancer — evidence for a relevant model system and urine-based diagnostic test. Mol. Cancer Res. 13, 993–1002 (2015).
Davies, H. et al. Mutations of the BRAF gene in human cancer. Nature 417, 949–954 (2002).
Puntervoll, H. E., Molven, A. & Akslen, L. A. Frequency of somatic BRAF mutations in melanocytic lesions from patients in a CDK4 melanoma family. Pigment Cell Melanoma Res. 27, 149–151 (2014).
DiMasi, J. A. & Grabowski, H. G. Economics of new oncology drug development. J. Clin. Oncol. 25, 209–216 (2007).
Nowinsky, M. Zur Frage ueber die Impfung der krebsigen Geschwuelste. Zentralbl Med. Wissensch 14, 790–791 (1876).
Murgia, C., Pritchard, J. K., Kim, S. Y., Fassati, A. & Weiss, R. A. Clonal origin and evolution of a transmissible cancer. Cell 126, 477–487 (2006).
Strakova, A. & Murchison, E. P. The changing global distribution and prevalence of canine transmissible venereal tumour. BMC Vet. Res. 10, 168 (2014).
Katzir, N., Arman, E., Cohen, D., Givol, D. & Rechavi, G. Common origin of transmissible venereal tumors (TVT) in dogs. Oncogene 1, 445–448 (1987).
Murchison, E. P. et al. Transmissible dog cancer genome reveals the origin and history of an ancient cell lineage. Science 343, 437–440 (2014).
Overall, K. L. Natural animal models of human psychiatric conditions: assessment of mechanism and validity. Prog. Neuropsychopharmacol. Biol. Psychiatry 24, 727–776 (2000).
Moon-Fanelli, A. A. & Dodman, N. H. Description and development of compulsive tail chasing in terriers and response to clomipramine treatment. J. Am. Vet. Med. Assoc. 212, 1252–1257 (1998).
Moon-Fanelli, A. A., Dodman, N. H. & Cottam, N. Blanket and flank sucking in Doberman Pinschers. J. Am. Vet. Med. Assoc. 231, 907–912 (2007).
Rapoport, J. L., Ryland, D. H. & Kriete, M. Drug treatment of canine acral lick. An animal model of obsessive-compulsive disorder. Arch. Gen. Psychiatry 49, 517–521 (1992).
Tang, R. et al. Candidate genes and functional noncoding variants identified in a canine model of obsessive-compulsive disorder. Genome Biol. 15, R25 (2014).
Dodman, N. H. et al. A canine chromosome 7 locus confers compulsive disorder susceptibility. Mol. Psychiatry 15, 8–10 (2010).
Nazaryan, L. et al. Association study between CDH2 and Gilles de la Tourette syndrome in a Danish cohort. Psychiatry Res. 228, 974–975 (2015).
Goodloe, L. P. & Borchelt, P. L. Companion dog temperament traits. J. Appl. Anim. Welf. Sci. 1, 303–338 (1998).
van den Berg, L., Schilder, M. B., de Vries, H., Leegwater, P. A. & van Oost, B. A. Phenotyping of aggressive behavior in golden retriever dogs with a questionnaire. Behav. Genet. 36, 882–902 (2006).
van den Berg, L., Schilder, M. B. & Knol, B. W. Behavior genetics of canine aggression: behavioral phenotyping of golden retrievers by means of an aggression test. Behav. Genet. 33, 469–483 (2003).
Hsu, Y. & Serpell, J. A. Development and validation of a questionnaire for measuring behavior and temperament traits in pet dogs. J. Am. Vet. Med. Assoc. 223, 1293–1300 (2003).
Zapata, I., Serpell, J. A. & Alvarez, C. E. Genetic mapping of canine fear and aggression. BMC Genomics 17, 572 (2016).
Saetre, P. et al. The genetic contribution to canine personality. Genes Brain Behav. 5, 240–248 (2006).
Schutt, T., Toft, N. & Berendt, M. Cognitive function, progression of age-related behavioral changes, biomarkers, and survival in dogs more than 8 years old. J. Vet. Intern. Med. 29, 1569–1577 (2015).
Fast, R., Schutt, T., Toft, N., Moller, A. & Berendt, M. An observational study with long-term follow-up of canine cognitive dysfunction: clinical characteristics, survival, and risk factors. J. Vet. Intern. Med. 27, 822–829 (2013).
Rofina, J. E. et al. Cognitive disturbances in old dogs suffering from the canine counterpart of Alzheimer's disease. Brain Res. 1069, 216–226 (2006).
Borghys, H. et al. Young to middle-aged dogs with high amyloid-beta levels in cerebrospinal fluid are impaired on learning in standard cognition tests. J. Alzheimers Dis. 56, 763–774 (2016).
Schutt, T. et al. Dogs with cognitive dysfunction as a spontaneous model for early alzheimer's disease: a translational study of neuropathological and inflammatory markers. J. Alzheimers Dis. 52, 433–449 (2016).
Bosch, M. N., Gimeno-Bayon, J., Rodriguez, M. J., Pugliese, M. & Mahy, N. Rapid improvement of canine cognitive dysfunction with immunotherapy designed for Alzheimer's disease. Curr. Alzheimer Res. 10, 482–493 (2013).
Neff, M. W. & Rine, J. A fetching model organism. Cell 124, 229–231 (2006).
Spady, T. C. & Ostrander, E. A. Canine behavioral genetics: pointing out the phenotypes and herding up the genes. Am. J. Hum. Genet. 82, 10–18 (2008).
American Kennel Club. Sporting Group. American Kennel Club http://www.akc.org/dog-breeds/groups/sporting/ (2017).
Chase, K., Jones, P., Martin, A., Ostrander, E. A. & Lark, K. G. Genetic mapping of fixed phenotypes: disease frequency as a breed characteristic. J. Hered. 100 (Suppl. 1), 37–41 (2009).
Akkad, D. A., Gerding, W. M., Gasser, R. B. & Epplen, J. T. Homozygosity mapping and sequencing identify two genes that might contribute to pointing behavior in hunting dogs. Canine Genet. Epidemiol. 2, 5 (2015).
vonHoldt, B. M. et al. Structural variants in genes associated with human Williams-Beuren syndrome underlie stereotypical hypersociability in domestic dogs. Sci. Adv. 3, e1700398 (2017).
Slatkin, M. & Racimo, F. Ancient DNA and human history. Proc. Natl Acad. Sci. USA 113, 6380–6387 (2016).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Barrangou, R. Diversity of CRISPR-Cas immune systems and molecular machines. Genome Biol. 16, 247 (2015).
Hendel, A. et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 33, 985–989 (2015).
Mali, P., Esvelt, K. M. & Church, G. M. Cas9 as a versatile tool for engineering biology. Nat. Methods 10, 957–963 (2013).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Zou, Q. et al. Generation of gene-target dogs using CRISPR/Cas9 system. J. Mol. Cell. Biol. 7, 580–583 (2015).
Mosher, D. S. et al. A mutation in the myostatin gene increases muscle mass and enhances racing performance in heterozygote dogs. PLoS Genet. 3, e79 (2007).
Karlsson, E. K. et al. Efficient mapping of mendelian traits in dogs through genome-wide association. Nat. Genet. 39, 1321–1328 (2007).
Whiteley, M. H., Bell, J. S. & Rothman, D. A. Novel allelic variants in the canine cyclooxgenase-2 (Cox-2) promoter are associated with renal dysplasia in dogs. PLoS ONE 6, e16684 (2011).
Plassais, J. et al. A point mutation in a lincRNA upstream of GDNF is associated to a canine insensitivity to pain: a spontaneous model for human sensory neuropathies. PLoS Genet. 12, e1006482 (2016).
Biesiadecki, B. J., Elder, B. D., Yu, Z. B. & Jin, J. P. Cardiac troponin T variants produced by aberrant splicing of multiple exons in animals with high instances of dilated cardiomyopathy. J. Biol. Chem. 277, 50275–50285 (2002).
Wang, G. D. et al. Genetic convergence in the adaptation of dogs and humans to the high-altitude environment of the tibetan plateau. Genome Biol. Evol. 6, 2122–2128 (2014).
Acknowledgements
We gratefully acknowledge the many individuals who provided comments and edits for this paper. We thank Dayna Dreger, Heidi Parker and Andrew Hogan for providing figures and valuable suggestions. E.A.O. and B.W.D. are supported by the Intramural Program of the US National Human Genome Research Institute. B.W.D. also acknowledges support from Texas A&M University. R.K.W. acknowledges support from the US National Science Foundation grants DEB 1021397 and 1257716 and seminal insights from John Novembre, Kirk Lohmueller and Claire Marsden, all of which have greatly enhanced this perspective on canine evolution.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Population bottlenecks
-
Reduction in the size of a population due to any of a variety of factors (for example, natural disasters, disease or human intervention) that in turn reduces genetic variation in the population.
- Haplotype
-
A group of variants or markers on a chromosome that are inherited together from one generation to the next. It can also refer to a pattern of variation observed across members of a population.
- Linkage disequilibrium
-
(LD). Nonrandom association of alleles located at distinct loci; measured by determining if the frequency of two loci co-occurring is higher than expected by chance.
- Selective sweep
-
A decrease in genomic variation surrounding a mutation due to positive selection for the mutation.
- Genetic drift
-
Allele frequency changes in a population due to random mating of members of the population.
- Effective population size
-
(Ne). The number of individuals that contribute equally to inherited genetic variation to the next generation within a given population.
- Population subdivision
-
The relational structure of species with multiple subpopulations that exist either in total isolation or with minimal gene flow between them.
- Admixture
-
The process by which isolated populations initiate previously non-existent gene flow.
- Popular-sire effect
-
A reduction in genetic diversity in a population due to nonrandom and excess mating of a sire with desirable traits.
- Genetic load
-
A reduction in the mean individual fitness of a population due to the presence of deleterious alleles or allelic combinations relative to a genotypically ideal population.
- Non-synonymous
-
A change in DNA sequence that alters the encoded amino acid, thus altering the encoded protein.
- Synonymous
-
A change in DNA sequence which, if it occurs in a coding region, does not alter the resultant amino acid.
- Chondrodysplastic
-
A state of abnormal cartilaginous growth resulting in disproportionate dwarfism. In dogs, this affects only the limbs, with minimal other observed effects.
- Identical-by-descent
-
A haplotype shared between individuals that is inherited from a recent common ancestor without intervening recombination.
- Quantitative trait locus
-
(QTL). A defined region of DNA that correlates with variation in a phenotype. Quantitative traits, by comparison, are phenotypes that vary in degree or presentation due to the joint effects of multiple genes.
- Penetrance
-
The proportion of individuals in a population who display a given phenotype in the presence of a specific genotype.
- Introgression
-
Gene flow from one population or individual into the gene pool of another by repeated crosses between related individuals, resulting in individuals with genetic components from both initial populations.
Rights and permissions
About this article
Cite this article
Ostrander, E., Wayne, R., Freedman, A. et al. Demographic history, selection and functional diversity of the canine genome. Nat Rev Genet 18, 705–720 (2017). https://doi.org/10.1038/nrg.2017.67
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg.2017.67
This article is cited by
-
Genetic dissection of behavioral traits related to successful training of drug detection dogs
Scientific Reports (2023)
-
Machine learning prediction and classification of behavioral selection in a canine olfactory detection program
Scientific Reports (2023)
-
Going beyond established model systems of Alzheimer’s disease: companion animals provide novel insights into the neurobiology of aging
Communications Biology (2023)
-
Anthropogenic hybridization and its influence on the adaptive potential of the Sardinian wild boar (Sus scrofa meridionalis)
Journal of Applied Genetics (2023)
-
Dog models of human atherosclerotic cardiovascular diseases
Mammalian Genome (2023)