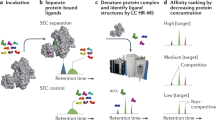
Abstract
NMR spectroscopy can be used to quantify the binding affinity between proteins and low-complexity molecules, termed 'fragments'; this versatile screening approach allows researchers to assess the druggability of new protein targets. Protein-observed 19F-NMR (PrOF NMR) using 19F-labeled amino acids generates relatively simple spectra that are able to provide dynamic structural information toward understanding protein folding and function. Changes in these spectra upon the addition of fragment molecules can be observed and quantified. This protocol describes the sequence-selective labeling of three proteins (the first bromodomains of Brd4 and BrdT, and the KIX domain of the CREB-binding protein) using commercially available fluorinated aromatic amino acids and fluorinated precursors as example applications of the method developed by our research group. Fragment-screening approaches are discussed, as well as Kd determination, ligand-efficiency calculations and druggability assessment, i.e., the ability to target these proteins using small-molecule ligands. Experiment times on the order of a few minutes and the simplicity of the NMR spectra obtained make this approach well-suited to the investigation of small- to medium-sized proteins, as well as the screening of multiple proteins in the same experiment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Lepre, C.A. Practical aspects of NMR-based fragment screening. Methods Enzymol. 493, 219–239 (2011).
Dalvit, C. et al. A General NMR method for rapid, efficient, and reliable biochemical screening. J. Am. Chem. Soc. 125, 14620–14625 (2003).
Fielding, L. NMR methods for the determination of protein-ligand dissociation constants. Curr. Top. Med. Chem. 3, 39–53 (2003).
Williamson, M.P. Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 73, 1–16 (2013).
Tanaka, D. et al. A Practical use of ligand efficiency indices out of the fragment-based approach: ligand efficiency-guided lead identification of soluble epoxide hydrolase inhibitors. J. Med. Chem. 54, 851–857 (2010).
Hopkins, A.L., Keseru, G.M., Leeson, P.D., Rees, D.C. & Reynolds, C.H. The role of ligand efficiency metrics in drug discovery. Nat. Rev. Drug Discov. 13, 105–121 (2014).
Shuker, S.B., Hajduk, P.J., Meadows, R.P. & Fesik, S.W. Discovering high-affinity ligands for proteins: SAR by NMR. Science 274, 1531–1534 (1996).
Erlanson, D.A., McDowell, R.S. & O'Brien, T. Fragment-based drug discovery. J. Med. Chem. 47, 3463–3482 (2004).
Bollag, G. et al. Vemurafenib: the first drug approved for BRAF-mutant cancer. Nat. Rev. Drug Discov. 11, 873–886 (2012).
Hajduk, P.J. & Greer, J. A decade of fragment-based drug design: strategic advances and lessons learned. Nat. Rev. Drug Discov. 6, 211–219 (2007).
Arntson, K.E. & Pomerantz, W.C.K. Protein-observed fluorine NMR: a bioorthogonal approach for small molecule discovery. J. Med. Chem. 59, 5158–5171 (2016).
Kitevski-LeBlanc, J.L. & Prosser, R.S. Current applications of 19F NMR to studies of protein structure and dynamics. Prog. Nucl. Magn. Reson. Spectrosc. 62, 1–33 (2012).
Sharaf, N.G. & Gronenborn, A.M. (19)F-Modified proteins and (19)F-containing ligands as tools in solution NMR studies of protein interactions. Methods Enzymol. 565, 67–95 (2015).
Gerig, J. Fluorine NMR. Biophysics Textbook Online 1–35 (2001).
Wu, B. et al. HTS by NMR of combinatorial libraries: a fragment-based approach to ligand discovery. Chem. Biol. 20, 19–33 (2013).
Doak, B.C., Morton, C.J., Simpson, J.S. & Scanlon, M.J. Design and evaluation of the performance of an NMR screening fragment library. Aust. J. Chem. 66, 1465–1472 (2013).
Siegal, G., Ab, E. & Schultz, J. Integration of fragment screening and library design. Drug Discov. Today 12, 1032–1039 (2007).
Lim, S.S. et al. Development of inhibitors of Plasmodium falciparum apical membrane antigen 1 based on fragment screening. Aust. J. Chem. 66, 1530 (2013).
Vom, A. et al. Detection and prevention of aggregation-based false positives in STD-NMR-based fragment screening. Aust. J. Chem. 66, 1518 (2013).
Dalvit, C., Fagerness, P.E., Hadden, D.T., Sarver, R.W. & Stockman, B.J. Fluorine-NMR experiments for high-throughput screening: theoretical aspects, practical considerations, and range of applicability. J. Am. Chem. Soc. 125, 7696–7703 (2003).
Dias, D.M. et al. Is NMR fragment screening fine-tuned to assess druggability of protein–protein interactions? ACS Med. Chem. Lett. 5, 23–28 (2013).
Pomerantz, W.C. et al. Profiling the dynamic interfaces of fluorinated transcription complexes for ligand discovery and characterization. ACS Chem. Biol. 7, 1345–1350 (2012).
Gee, C.T., Koleski, E.J. & Pomerantz, W.C. Fragment screening and druggability assessment for the CBP/p300 KIX domain through protein-observed 19F NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 54, 3735–3739 (2015).
Leung, E.W. et al. 19F NMR as a probe of ligand interactions with the iNOS binding site of SPRY domain-containing SOCS box protein 2. Chem. Biol. Drug Des. 84, 616–625 (2014).
Ge, X. et al. Ligand-induced conformational change of Plasmodium falciparum AMA1 detected using 19F NMR. J. Med. Chem. 57, 6419–6427 (2014).
Curtis-Marof, R. et al. 19F NMR spectroscopy monitors ligand binding to recombinantly fluorine-labelled b'x from human protein disulphide isomerase (hPDI). Org. Biomol. Chem. 12, 3808–3812 (2014).
Mishra, N.K., Urick, A.K., Ember, S.W., Schonbrunn, E. & Pomerantz, W.C. Fluorinated aromatic amino acids are sensitive 19F NMR probes for bromodomain-ligand interactions. ACS Chem. Biol. 9, 2755–2760 (2014).
Liu, J.J., Horst, R., Katritch, V., Stevens, R.C. & Wuthrich, K. Biased signaling pathways in beta(2)-adrenergic receptor characterized by F-19-NMR. Science 335, 1106–1110 (2012).
Yu, L., Hajduk, P.J., Mack, J. & Olejniczak, E.T. Structural studies of Bcl-xL/ligand complexes using 19F NMR. J. Biomol. NMR 34, 221–227 (2006).
Urick, A.K. et al. Dual screening of BPTF and Brd4 using protein-observed fluorine NMR Uncovers new bromodomain probe molecules. ACS Chem. Biol. 10 2246–2256 (2015).
Zartler, E.R. et al. RAMPED-UP NMR: multiplexed NMR-based screening for drug discovery. J. Am. Chem. Soc. 125, 10941–10946 (2003).
Sykes, B.D., Weingarten, H.I. & Schlesinger, M.J. Fluorotyrosine alkaline phosphatase from Escherichia coli: preparation, properties, and fluorine-19 nuclear magnetic resonance spectrum. Proc. Natl. Acad. Sci. USA 71, 469–473 (1974).
Frieden, C., Hoeltzli, S.D. & Bann, J.G. The preparation of 19F-labeled proteins for NMR studies. in Methods Enzymol. Vol. 380 (eds. Michael, L., Johnson Jo M. Holt & K. Ackers Gary) 400–415 (Academic Press, 2004).
Crowley, P.B., Kyne, C. & Monteith, W.B. Simple and inexpensive incorporation of 19F-tryptophan for protein NMR spectroscopy. Chem. Commun. 48, 10681–10683 (2012).
Martin, M.P., Olesen, S.H., Georg, G.I. & Schönbrunn, E. Cyclin-dependent kinase inhibitor dinaciclib interacts with the acetyl-lysine recognition site of bromodomains. ACS Chem. Biol. 8, 2360–2365 (2013).
Bryant, R.G. The NMR time scale. J. Chem. Educ. 60, 933 (1983).
Rogers, M.T. & Woodbrey, J.C. A proton magnetic resonance study of hindered internal rotation in some substituted N,N-dimethylamides. J. Phys. Chem. 66, 540–546 (1962).
Hull, W.E. & Sykes, B.D. Fluorotyrosine alkaline phosphatase: internal mobility of individual tyrosines and the role of chemical shift anisotropy as a 19F nuclear spin relaxation mechanism in proteins. J. Mol. Biol. 98, 121–153 (1975).
Ho, C., Pratt, E.A. & Rule, G.S. Membrane-bound D-lactate dehydrogenase of Escherichia coli: a model for protein interactions in membranes. Biochim. Biophys. Acta 988, 173–184 (1989).
Klein-Seetharaman, J., Getmanova, E.V., Loewen, M.C., Reeves, P.J. & Khorana, H.G. NMR spectroscopy in studies of light-induced structural changes in mammalian rhodopsin: applicability of solution 19F NMR. Proc. Natl. Acad. Sci. USA 96, 13744–13749 (1999).
Chung, K.Y. et al. Role of detergents in conformational exchange of a G protein-coupled receptor. J. Biol. Chem. 287, 36305–36311 (2012).
Suzuki, Y. et al. Resolution of oligomeric species during the aggregation of Abeta1–40 using (19)F NMR. Biochemistry 52, 1903–1912 (2013).
Li, C. et al. Protein (19)F NMR in Escherichia coli. J. Am. Chem. Soc. 132, 321 (2010).
Hammill, J.T., Miyake-Stoner, S., Hazen, J.L., Jackson, J.C. & Mehl, R.A. Preparation of site-specifically labeled fluorinated proteins for 19F-NMR structural characterization. Nat. Protoc. 2, 2601–2607 (2007).
Salopek-Sondi, B., Vaughan, M.D., Skeels, M.C., Honek, J.F. & Luck, L.A. (19)F NMR studies of the leucine-isoleucine-valine binding protein: evidence that a closed conformation exists in solution. J. Biomol. Struct. Dyn. 21, 235–246 (2003).
Duewel, H., Daub, E., Robinson, V. & Honek, J.F. Incorporation of trifluoromethionine into a phage lysozyme: implications and a new marker for use in protein 19F NMR. Biochemistry 36, 3404–3416 (1997).
Tang, Y. & Tirrell, D.A. Biosynthesis of a highly stable coiled-coil protein containing hexafluoroleucine in an engineered bacterial host. J. Am. Chem. Soc. 123, 11089–11090 (2001).
Tang, Y. et al. Stabilization of coiled-coil peptide domains by introduction of trifluoroleucine. Biochemistry 40, 2790–2796 (2001).
Lee, H.Y., Lee, K.H., Al-Hashimi, H.M. & Marsh, E.N. Modulating protein structure with fluorous amino acids: increased stability and native-like structure conferred on a 4-helix bundle protein by hexafluoroleucine. J. Am. Chem. Soc. 128, 337–343 (2006).
Bogan, A.A. & Thorn, K.S. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 280, 1–9 (1998).
Hajduk, P.J., Huth, J.R. & Fesik, S.W. Druggability indices for protein targets derived from NMR-based screening data. J. Med. Chem. 48, 2518–2525 (2005).
Lodge, J.M., Justin Rettenmaier, T., Wells, J.A., Pomerantz, W.C. & Mapp, A.K. FP tethering: a screening technique to rapidly identify compounds that disrupt protein–protein interactions. Medchemcomm. 5, 370–375 (2014).
Kim, H.W., Perez, J.A., Ferguson, S.J. & Campbell, I.D. The specific incorporation of labelled aromatic amino acids into proteins through growth of bacteria in the presence of glyphosate. Application to fluorotryptophan labelling to the H(+)-ATPase of Escherichia coli and NMR studies. FEBS Lett. 272, 34–36 (1990).
Bai, P., Luo, L. & Peng, Z. Side chain accessibility and dynamics in the molten globule state of alpha-lactalbumin: a (19)F-NMR study. Biochemistry 39, 372–380 (2000).
Neerathilingam, M., Greene, L.H., Colebrooke, S.A., Campbell, I.D. & Staunton, D. Quantitation of protein expression in a cell-free system: efficient detection of yields and 19F NMR to identify folded protein. J. Biomol. NMR 31, 11–19 (2005).
Akoka, S., Barantin, L. & Trierweiler, M. Concentration measurement by proton NMR using the ERETIC method. Anal. Chem. 71, 2554–2557 (1999).
Dalvit, C. et al. Sensitivity improvement in 19F NMR-based screening experiments: theoretical considerations and experimental applications. J. Am. Chem. Soc. 127, 13380–13385 (2005).
Seyedsayamdost, M.R., Reece, S.Y., Nocera, D.G. & Stubbe, J. Mono-, di-, tri-, and tetra-substituted fluorotyrosines: new probes for enzymes that use tyrosyl radicals in catalysis. J. Am. Chem. Soc. 128, 1569–1579 (2006).
Furter, R. Expansion of the genetic code: site-directed p-fluoro-phenylalanine incorporation in Escherichia coli. Prot. Sci. 7, 419–426 (1998).
Kitevski-LeBlanc, J.L., Al-Abdul-Wahid, M.S. & Prosser, R.S. A mutagenesis-free approach to assignment of 19F NMR resonances in biosynthetically labeled proteins. J. Am. Chem. Soc. 131, 2054–2055 (2009).
Acknowledgements
This project was funded in part by the NSF-CAREER Award CHE-1352091 (to W.C.K.P., N.K.M. and L.M.L.H.), National Institutes of Health (NIH) Biotechnology training grant 5T32GM008347-23 (to A.K.U.) and NIH chemistry–biology interface training grant T32-GM08700 (to C.T.G.). The Pomerantz lab also thanks the Garber family and relatives for their generous support of this research. We also thank I. Ropson (Penn State University) for providing the DL39(DE3) cell line.
Author information
Authors and Affiliations
Contributions
W.C.K.P. oversaw the implementation and design of all experiments reported here. C.T.G. and K.E.A. carried out protein expression and characterization experiments with KIX; C.T.G. also carried out protein expression and characterization experiments with BrdT; N.K.M. and A.K.U. carried out protein expression and characterization experiments with Brd4; L.M.L.H. carried out fluorescence polarization experiments; and A.W. discovered fragment A. All authors assisted with writing and editing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Binding activity comparison of non-fluorinated and fluorinated proteins
Direct binding experiment of BODIPY-BI, a Brd4 bromodomain ligand to unlabeled Brd4 and 5FW-labeled Brd4 by fluorescence polarization.
Supplementary Figure 2 19F NMR spectral analysis of 5FW BrdT in the presence of increasing concentrations of Dinaciclib.
A) Left: Ribbon diagram of BrdT (PDB Code: 4FLP) with tryptophan side chains indicated as sticks shown in red. Right: Stacked PrOF NMR spectra with increasing concentration of dinaciclib. B) Absolute value of chemical shift perturbations for all 5FW BrdT tyrosine resonances at 400 μM Dinaciclib. C) Binding isotherm of both W44* and W50* perturbations for the titration with Dinaciclib. * denotes resonances that have not yet been assigned by site directed mutagenesis, but were inferred from ligand binding.
Supplementary Figure 3 19F NMR spectral analysis of 5FW BrdT in the presence of increasing concentrations of A.
A) Left: Ribbon diagram of BrdT (PDB Code: 4FLP) with tryptophan side chains indicated as sticks shown in red. Right: Stacked PrOF NMR spectra with increasing concentration of A. B) Absolute value of chemical shift perturbations for all 5FW BrdT tyrosine resonances at 2 mM A. C) Binding isotherm of both W44* and W50* perturbations for the titration with A. * denotes resonances that have not yet been assigned by site directed mutagenesis, but were inferred from ligand binding.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3, Supplementary Method, Supplementary Tables 1 and 2 (PDF 1604 kb)
Rights and permissions
About this article
Cite this article
Gee, C., Arntson, K., Urick, A. et al. Protein-observed 19F-NMR for fragment screening, affinity quantification and druggability assessment. Nat Protoc 11, 1414–1427 (2016). https://doi.org/10.1038/nprot.2016.079
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.079
This article is cited by
-
2-Fluorotyrosine is a valuable but understudied amino acid for protein-observed 19F NMR
Journal of Biomolecular NMR (2020)
-
Two-dimensional 19F–13C correlation NMR for 19F resonance assignment of fluorinated proteins
Journal of Biomolecular NMR (2020)
-
Aromatic 19F-13C TROSY: a background-free approach to probe biomolecular structure, function, and dynamics
Nature Methods (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.