Abstract
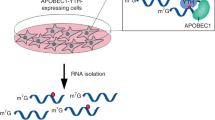
N6-methyladenosine–sequencing (m6A-seq) is an immunocapturing approach for the unbiased transcriptome-wide localization of m6A in high resolution. To our knowledge, this is the first protocol to allow a global view of this ubiquitous RNA modification, and it is based on antibody-mediated enrichment of methylated RNA fragments followed by massively parallel sequencing. Building on principles of chromatin immunoprecipitation–sequencing (ChIP-seq) and methylated DNA immunoprecipitation (MeDIP), read densities of immunoprecipitated RNA relative to untreated input control are used to identify methylated sites. A consensus motif is deduced, and its distance to the point of maximal enrichment is assessed; these measures further corroborate the success of the protocol. Identified locations are intersected in turn with gene architecture to draw conclusions regarding the distribution of m6A between and within gene transcripts. When applied to human and mouse transcriptomes, m6A-seq generated comprehensive methylation profiles revealing, for the first time, tenets governing the nonrandom distribution of m6A. The protocol can be completed within ∼9 d for four different sample pairs (each consists of an immunoprecipitation and corresponding input).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Cantara, W.A. et al. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res. 39, D195–D201 (2011).
He, C. Grand challenge commentary: RNA epigenetics? Nat. Chem. Biol. 6, 863–865 (2010).
Chan, C.T. et al. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 6, e1001247 (2010).
Schaefer, M. et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 24, 1590–1595 (2010).
Bokar, J. Fine-tuning of RNA functions by modification and editing. in Topics in Current Genetics 12 (ed. Grosjean, H.) 141–177 (Springer, 2005).
Zhong, S. et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 20, 1278–1288 (2008).
Clancy, M.J., Shambaugh, M.E., Timpte, C.S. & Bokar, J.A. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 30, 4509–4518 (2002).
Jia, G. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 (2011).
Levanon, E.Y. et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 22, 1001–1005 (2004).
Klose, R.J. & Bird, A.P. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 31, 89–97 (2006).
Dai, Q. et al. Identification of recognition residues for ligation-based detection and quantitation of pseudouridine and N6-methyladenosine. Nucleic Acids Res. 35, 6322–6329 (2007).
Kellner, S., Burhenne, J. & Helm, M. Detection of RNA modifications. RNA biology 7, 237–247 (2010).
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012).
Meyer, K.D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012).
Czerwoniec, A. et al. MODOMICS: a database of RNA modification pathways. 2008 update. Nucleic Acids Res. 37, D118–121 (2009).
Horowitz, S., Horowitz, A., Nilsen, T.W., Munns, T.W. & Rottman, F.M. Mapping of N6-methyladenosine residues in bovine prolactin mRNA. Proc. Natl. Acad. Sci. USA 81, 5667–5671 (1984).
Bringmann, P. & Luhrmann, R. Antibodies specific for N6-methyladenosine react with intact snRNPs U2 and U4/U6. FEBS Lett. 213, 309–315 (1987).
Dante, R. & Niveleau, A. Inhibition of in vitro translation by antibodies directed against N6-methyladenosine. FEBS Lett. 130, 153–157 (1981).
Munns, T.W., Liszewski, M.K., Oberst, R.J. & Sims, H.F. Antibody nucleic acid complexes. Immunospecific retention of N6-methyladenosine-containing transfer ribonucleic acid. Biochemistry 17, 2573–2578 (1978).
Munns, T.W., Liszewski, M.K. & Sims, H.F. Characterization of antibodies specific for N6-methyladenosine and for 7-methylguanosine. Biochemistry 16, 2163–2168 (1977).
Munns, T.W., Oberst, R.J., Sims, H.F. & Liszewski, M.K. Antibody-nucleic acid complexes. Immunospecific recognition of 7-methylguanine- and N6-methyladenine-containing 5′-terminal oligonucleotides of mRNA. J. Biol. Chem. 254, 4327–4330 (1979).
Munns, T.W., Sims, H.F. & Liszewski, M.K. Immunospecific retention of oligonucleotides possessing N6-methyladenosine and 7-methylguanosine. J. Biol. Chem. 252, 3102–3104 (1977).
Zhang, Y. et al. Model-based analysis of ChIP-seq (MACS). Genome Biol. 9, R137 (2008).
Feng, J., Liu, T. & Zhang, Y. Using MACS to identify peaks from ChIP-seq data. Curr. Protoc. Bioinformatics 34, 2.14.1–2.14.14 (2011).
Machanick, P. & Bailey, T.L. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27, 1696–1697 (2011).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Quinlan, A.R. & Hall, I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Bailey, T.L. & Machanick, P. Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res. 18, 18 (2012).
Salmon-Divon, M., Dvinge, H., Tammoja, K. & Bertone, P. PeakAnalyzer: genome-wide annotation of chromatin binding and modification loci. BMC Bioinformatics 11, 415 (2010).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010).
Trapnell, C., Pachter, L. & Salzberg, S.L. TopHat: discovering splice junctions with RNA-seq. Bioinformatics 25, 1105–1111 (2009).
Langmead, B. & Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Acknowledgements
We thank the Kahn Family Foundation for their support. This work was supported in part by grants from the Flight Attendant Medical Research Institute (FAMRI), Bio-Med Morasha Israel Science Foundation (ISF) (grant no. 1942/08), ISF (grant no. 1667/12), the molecular basis of human disease I-CORE (Israeli Centers of Research Excellence) and the Israel Ministry of Science and Technology (Scientific Infrastructure Program). G.R. holds the Djerassi Chair in Oncology at the Sackler Faculty of Medicine, Tel Aviv University. This work was performed in partial fulfillment of the requirements for a PhD degree to D.D., Sackler Faculty of Medicine, Tel Aviv University.
Author information
Authors and Affiliations
Contributions
D.D. and S.M.-M. conceived the approach, developed the protocol and performed the experiments. M.S.-D. designed the bioinformatic pipeline and analyzed the data. N.A. and G.R. supervised the project. D.D., S.M.-M., M.S.-D., N.A. and G.R. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Dominissini, D., Moshitch-Moshkovitz, S., Salmon-Divon, M. et al. Transcriptome-wide mapping of N6-methyladenosine by m6A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc 8, 176–189 (2013). https://doi.org/10.1038/nprot.2012.148
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2012.148
This article is cited by
-
The role of the methyltransferase METTL3 in prostate cancer: a potential therapeutic target
BMC Cancer (2024)
-
Circular RNA-circPan3 attenuates cardiac hypertrophy via miR-320-3p/HSP20 axis
Cellular & Molecular Biology Letters (2024)
-
New horizons for the role of RNA N6-methyladenosine modification in hepatocellular carcinoma
Acta Pharmacologica Sinica (2024)
-
The PIN1-YTHDF1 axis promotes breast tumorigenesis via the m6A-dependent stabilization of AURKA mRNA
Archives of Pharmacal Research (2024)
-
A chromatin-regulated biphasic circuit coordinates IL-1β-mediated inflammation
Nature Genetics (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.