Abstract
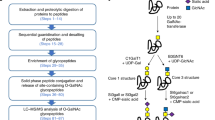
Protein post-translational modifications (PTMs), such as glycosylation and phosphorylation, are crucial for various signaling and regulatory events, and are therefore an important objective of proteomics research. We describe here a protocol for isotope-coded glycosylation site–specific tagging (IGOT), a method for the large-scale identification of N-linked glycoproteins from complex biological samples. The steps of this approach are: (1) lectin column–mediated affinity capture of glycopeptides generated by protease digestion of protein mixtures; (2) purification of the enriched glycopeptides by hydrophilic interaction chromatography (HIC); (3) peptide-N-glycanase-mediated incorporation of a stable isotope tag, 18O18O, specifically at the N-glycosylation site; and (4) identification of 18O-tagged peptides by liquid chromatography–coupled mass spectrometry (LC/MS)-based proteomics technology. The application of this protocol to the characterization of N-linked glycoproteins from crude extracts of the nematode Caenorhabditis elegans or mouse liver provides a list of hundreds to a thousand glycoproteins and their sites of glycosylation within a week.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Mann, M. & Jensen, O.N. Proteomic analysis of post-translational modifications. Nat. Biotechnol. 21, 255–261 (2003).
Krishna, R.G. & Wold, F. Post-translational modification of proteins. Adv. Enzymol. Relat. Areas Mol. Biol. 67, 265–298 (1993).
Freeze, H.H. & Aebi, M. Altered glycan structures: the molecular basis of congenital disorders of glycosylation. Curr. Opin. Struct. Biol. 15, 490–498 (2005).
Sturiale, L. et al. Hypoglycosylation with increased fucosylation and branching of serum transferrin N-glycans in untreated galactosemia. Glycobiology 15, 1268–1276 (2005).
Varki, A. et al. in Essentials of Glycobiology (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1999).
Petrescu, A.-J. et al. Statistical analysis of the protein environment of N-glycosylation sites: implications for occupancy, structure, and folding. Glycobiology 14, 103–114 (2004).
Jones, J., Krag, S.S. & Betenbaugh, M.J. Controlling N-linked glycan site occupancy. Biochim. Biophys. Acta 1726, 121–137 (2005).
Moloney, D.J. et al. Fringe is a glycosyltransferase that modifies Notch. Nature 406, 369–375 (2000).
Block, T.M. et al. Use of targeted glycoproteomics to identify serum glycoproteins that correlate with liver cancer in woodchucks and humans. Proc. Natl. Acad. Sci. USA 102, 779–784 (2005).
Hirabayashi, J. et al. Affinity capturing and gene assignment of soluble glycoproteins produced by the nematode Caenorhabditis elegans. J. Biochem. 132, 103–114 (2002).
Gonzalez, J. et al. A method for determination of N-glycosylation sites in glycoproteins by collision-induced dissociation analysis in fast atom bombardment mass spectrometry: identification of the positions of carbohydrate-linked asparagine in recombinant alpha-amylase by treatment with peptide-N-glycosidase F in 18O-labeled water. Anal. Biochem. 205, 151–158 (1992).
Zhang, H., Li, X.J., Martin, D.B. & Aebersold, R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol. 21, 660–666 (2003).
Kaji, H. et al. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat. Biotechnol. 21, 667–672 (2003).
Kameyama, A. et al. Strategy for simulation of CID spectra of N-linked oligosaccharides toward glycomics. J. Proteome Res. 5, 808–814 (2006).
Wada, Y., Tajiri, M. & Yoshida, S. Hydrophilic affinity isolation and MALDI multiple-stage tandem mass spectrometry of glycopeptides for glycoproteomics. Anal. Chem. 76, 6560–6565 (2004).
Cieniewski-Bernard, C. et al. Identification of O-linked N-acetylglucosamine proteins in rat skeletal muscle using two-dimensional gel electrophoresis and mass spectrometry. Mol. Cell. Proteomics 3, 577–585 (2004).
Vosseller, K. et al. O-linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol. Cell. Proteomics 5, 923–934 (2006).
Vocadlo, D.J., Hang, H.C., Kim, E.J., Hanover, J.A. & Bertozzi, C.R. A chemical approach for identifying O-GlcNAc modified proteins in cells. Proc. Natl. Acad. Sci. USA 100, 9116–9121 (2003).
Sprung, R. et al. Tagging-via-substrate strategy for probing O-GlcNAc modified proteins. J. Proteome Res. 4, 950–957 (2005).
Khidekel, N., Ficarro, S.B., Peters, E.C. & Hsieh-Wilson, L.C. Exploring the O-GlcNAc proteome: direct identification of O-GlcNAc-modified proteins from the brain. Proc. Natl. Acad. Sci. USA 101, 13132–13137 (2004).
Wells, L. et al. Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol. Cell. Proteomics 1, 791–804 (2002).
Oda, Y. et al. Accurate quantitation of protein expression and site-specific phosphorylation. Proc. Natl. Acad. Sci. USA 96, 6591–6596 (1999).
Ong, S.E. et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 (2002).
Shiio, Y. & Aebersold, R. Quantitative proteome analysis using isotope-coded affinity tags and mass spectrometry. Nat. Protocols 1, 139–145 (2006).
Natsume, T. et al. A direct nanoflow liquid chromatography-tandem mass spectrometry system for interaction proteomics. Anal. Chem. 74, 4725–4733 (2002).
Isobe, T. et al. Automated two-dimensional liquid chromatography/tandem mass spectrometry for large-scale protein analysis. in Proteins and Proteomics 869–876 (Cold Spring Harbor Press, Cold Spring Harbor, NY, 2003).
Mawuenyega, K. et al. Large-scale identification of Caenorhabditis elegans proteins by multi-dimensional liquid chromatography-tandem mass spectrometry. J. Proteome Res. 2, 23–35 (2003).
Takahashi, N. et al. Proteomic snapshot analyses of preribosomal ribonucleoprotein complexes formed at various stages of ribosome biogenesis in yeast and mammalian cells. Mass Spectrom. Rev. 22, 287–317 (2003).
Taoka, M. et al. Only a small subset of horizontally transferred chromosomal genes in Escherichia coli are expressed into proteins. Mol. Cell. Proteomics 3, 780–787 (2004).
Nagano, K. et al. Large-scale identification of proteins expressed in mouse embryonic stem cells. Proteomics 5, 1346–1361 (2005).
Nunomura, K. et al. Cell surface labeling and mass spectrometry reveal diversity of cell-surface markers and signaling molecules expressed in undifferentiated mouse embryonic stem cells. Mol. Cell. Proteomics 4, 1968–1976 (2005).
Shinkawa, T. et al. STEM: a software tool for large-scale proteomic data analyses. J. Proteome Res. 4, 1826–1831 (2005).
Ishida, Y., Fujita, T. & Asai, K. New detection and separation method for amino acids by high-performance liquid chromatography. J. Chromatogr. 204, 143–148 (1981).
Acknowledgements
This work was supported in part by grants for the Structural Glycomics Project from the New Energy and Industrial Technology Development Organization (NEDO) of Japan, and for the Integrated Proteomics Project, Pioneer Research on Genome the Frontier from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan, and by a grant-in-aid for Scientific Research from MEXT of Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Kaji, H., Yamauchi, Y., Takahashi, N. et al. Mass spectrometric identification of N-linked glycopeptides using lectin-mediated affinity capture and glycosylation site–specific stable isotope tagging. Nat Protoc 1, 3019–3027 (2006). https://doi.org/10.1038/nprot.2006.444
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2006.444
This article is cited by
-
Wisteria floribunda agglutinin staining for the quantitative assessment of cardiac fibrogenic activity in a mouse model of dilated cardiomyopathy
Laboratory Investigation (2019)
-
Recent advances in enhanced enzyme activity, thermostability and secretion by N-glycosylation regulation in yeast
Biotechnology Letters (2018)
-
Identification of Poly-N-Acetyllactosamine-Carrying Glycoproteins from HL-60 Human Promyelocytic Leukemia Cells Using a Site-Specific Glycome Analysis Method, Glyco-RIDGE
Journal of the American Society for Mass Spectrometry (2018)
-
Isolation, characterization and analysis of the agglutinative activity of a lectin from Crotalaria spectabilis
Journal of Plant Biochemistry and Biotechnology (2018)
-
Serum glycopattern and Maackia amurensis lectin-II binding glycoproteins in autism spectrum disorder
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.