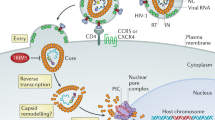
Abstract
To replicate in their hosts, viruses have to navigate the complexities of the mammalian cell, co-opting mechanisms of cellular physiology while defeating restriction factors that are dedicated to halting their progression. Primate lentiviruses devote a relatively large portion of their coding capacity to counteracting restriction factors by encoding accessory proteins dedicated to neutralizing the antiviral function of these intracellular inhibitors. Research into the roles of the accessory proteins has revealed the existence of previously undetected intrinsic defenses, provided insight into the evolution of primate lentiviruses as they adapt to new species and uncovered new targets for the development of therapeutics. This Review discusses the biology of the restriction factors APOBEC3, SAMHD1 and tetherin and the viral accessory proteins that counteract them.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Strebel, K. et al. The HIV 'A' (sor) gene product is essential for virus infectivity. Nature 328, 728–730 (1987).
Gabuzda, D.H. et al. Role of Vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J. Virol. 66, 6489–6495 (1992).
Cullen, B.R. HIV-1 Vif: counteracting innate antiretroviral defenses. Mol. Ther. 8, 525–527 (2003).
Simon, J.H., Gaddis, N.C., Fouchier, R.A. & Malim, M.H. Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat. Med. 4, 1397–1400 (1998).
Sheehy, A.M., Gaddis, N., Choi, J. & Malim, M. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418, 646–650 (2002).
Harris, R.S., Petersen-Mahrt, S.K. & Neuberger, M.S. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol. Cell 10, 1247–1253 (2002).
Jarmuz, A. et al. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79, 285–296 (2002).
Sawyer, S.L., Emerman, M. & Malik, H.S. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2, E275 (2004).
Conticello, S.G., Thomas, C.J., Petersen-Mahrt, S.K. & Neuberger, M.S. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol. Biol. Evol. 22, 367–377 (2005).
Refsland, E.W. et al. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. 38, 4274–4284 (2010).
Refsland, E.W. & Harris, R.S. The APOBEC3 family of retroelement restriction factors. Curr. Top. Microbiol. Immunol. 371, 1–27 (2013).
Desimmie, B.A. et al. Multiple APOBEC3 restriction factors for HIV-1 and one Vif to rule them all. J. Mol. Biol. 426, 1220–1245 (2014).
Mariani, R. et al. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114, 21–31 (2003).
Mangeat, B. et al. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424, 99–103 (2003).
Bishop, K.N. et al. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14, 1392–1396 (2004).
Apolonia, L. et al. Promiscuous RNA binding ensures effective encapsidation of APOBEC3 proteins by HIV-1. PLoS Pathog. 11, e1004609 (2015).
Zennou, V., Perez-Caballero, D., Gottlinger, H. & Bieniasz, P.D. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J. Virol. 78, 12058–12061 (2004).
Aguiar, R.S., Lovsin, N., Tanuri, A. & Peterlin, B.M. Vpr.A3A chimera inhibits HIV replication. J. Biol. Chem. 283, 2518–2525 (2008).
Suspène, R., Rusniok, C., Vartanian, J.P. & Wain-Hobson, S. Twin gradients in APOBEC3 edited HIV-1 DNA reflect the dynamics of lentiviral replication. Nucleic Acids Res. 34, 4677–4684 (2006).
Yu, Q. et al. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 11, 435–442 (2004).
Kim, E.Y. et al. Human APOBEC3 induced mutation of human immunodeficiency virus type-1 contributes to adaptation and evolution in natural infection. PLoS Pathog. 10, e1004281 (2014).
Holmes, R.K., Malim, M.H. & Bishop, K.N. APOBEC-mediated viral restriction: not simply editing? Trends Biochem. Sci. 32, 118–128 (2007).
Bishop, K.N., Verma, M., Kim, E.Y., Wolinsky, S.M. & Malim, M.H. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 4, e1000231 (2008).
Browne, E.P., Allers, C. & Landau, N.R. Restriction of HIV-1 by APOBEC3G is cytidine deaminase-dependent. Virology 387, 313–321 (2009).
Albin, J.S., Brown, W.L. & Harris, R.S. Catalytic activity of APOBEC3F is required for efficient restriction of Vif-deficient human immunodeficiency virus. Virology 450–451, 49–54 (2014).
Yu, X. et al. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302, 1056–1060 (2003).
Mehle, A., Thomas, E.R., Rajendran, K.S. & Gabuzda, D. A zinc-binding region in Vif binds Cul5 and determines cullin selection. J. Biol. Chem. 281, 17259–17265 (2006).
Jäger, S. et al. Vif hijacks CBF-β to degrade APOBEC3G and promote HIV-1 infection. Nature 481, 371–375 (2012).
Hultquist, J.F., Binka, M., LaRue, R.S., Simon, V. & Harris, R.S. Vif proteins of human and simian immunodeficiency viruses require cellular CBFbeta to degrade APOBEC3 restriction factors. J. Virol. 86, 2874–2877 (2012).
Kim, D.Y. et al. CBFbeta stabilizes HIV Vif to counteract APOBEC3 at the expense of RUNX1 target gene expression. Mol. Cell 49, 632–644 (2013).
Zhang, W., Du, J., Evans, S.L., Yu, Y. & Yu, X.F. T-cell differentiation factor CBF-β regulates HIV-1 Vif-mediated evasion of host restriction. Nature 481, 376–379 (2012).
Sheehy, A.M., Gaddis, N.C. & Malim, M.H. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9, 1404–1407 (2003).
Guo, Y. et al. Structural basis for hijacking CBF-beta and CUL5 E3 ligase complex by HIV-1 Vif. Nature 505, 229–233 (2014).
Simon, V. et al. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 1, e6 (2005).
Russell, R.A. & Pathak, V.K. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J. Virol. 81, 8201–8210 (2007).
Ooms, M. et al. HIV-1 Vif adaptation to human APOBEC3H haplotypes. Cell Host Microbe 14, 411–421 (2013).
Mulder, L.C., Harari, A. & Simon, V. Cytidine deamination induced HIV-1 drug resistance. Proc. Natl. Acad. Sci. USA 105, 5501–5506 (2008).
Kim, E.Y. et al. Human APOBEC3G-mediated editing can promote HIV-1 sequence diversification and accelerate adaptation to selective pressure. J. Virol. 84, 10402–10405 (2010).
Fourati, S. et al. Partially active HIV-1 Vif alleles facilitate viral escape from specific antiretrovirals. AIDS 24, 2313–2321 (2010).
Sato, K. et al. APOBEC3D and APOBEC3F potently promote HIV-1 diversification and evolution in humanized mouse model. PLoS Pathog. 10, e1004453 (2014).
Krisko, J.F., Martinez-Torres, F., Foster, J.L. & Garcia, J.V. HIV restriction by APOBEC3 in humanized mice. PLoS Pathog. 9, e1003242 (2013).
Casartelli, N. et al. The antiviral factor APOBEC3G improves CTL recognition of cultured HIV-infected T cells. J. Exp. Med. 207, 39–49 (2010).
Iwabu, Y., Fujita, H., Tanaka, Y., Sata, T. & Tokunaga, K. Direct internalization of cell-surface BST-2/tetherin by the HIV-1 accessory protein Vpu. Commun. Integr. Biol. 3, 366–369 (2010).
Iwabu, Y. et al. Differential anti-APOBEC3G activity of HIV-1 Vif proteins derived from different subtypes. J. Biol. Chem. 285, 35350–35358 (2010).
Binka, M., Ooms, M., Steward, M. & Simon, V. The activity spectrum of Vif from multiple HIV-1 subtypes against APOBEC3G, APOBEC3F, and APOBEC3H. J. Virol. 86, 49–59 (2012).
OhAinle, M., Kerns, J.A., Li, M.M.H., Malik, H.S. & Emerman, M. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe 4, 249–259 (2008).
Larue, R.S., Lengyel, J., Jonsson, S.R., Andresdottir, V. & Harris, R.S. Lentiviral Vif degrades the APOBEC3Z3/APOBEC3H protein of its mammalian host and is capable of cross-species activity. J. Virol. 84, 8193–8201 (2010).
Tan, L., Sarkis, P.T., Wang, T., Tian, C. & Yu, X.F. Sole copy of Z2-type human cytidine deaminase APOBEC3H has inhibitory activity against retrotransposons and HIV-1. FASEB J. 23, 279–287 (2009).
Harari, A., Ooms, M., Mulder, L.C. & Simon, V. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J. Virol. 83, 295–303 (2009).
Refsland, E.W. et al. Natural polymorphisms in human APOBEC3H and HIV-1 Vif combine in primary T lymphocytes to affect viral G-to-A mutation levels and infectivity. PLoS Genet. 10, e1004761 (2014).
Bogerd, H.P., Doehle, B.P., Wiegand, H.L. & Cullen, B.R. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc. Natl. Acad. Sci. USA 101, 3770–3774 (2004).
Mangeat, B., Turelli, P., Liao, S. & Trono, D. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J. Biol. Chem. 279, 14481–14483 (2004).
Schröfelbauer, B., Chen, D. & Landau, N.R. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc. Natl. Acad. Sci. USA 101, 3927–3932 (2004).
Krupp, A. et al. APOBEC3G polymorphism as a selective barrier to cross-species transmission and emergence of pathogenic SIV and AIDS in a primate host. PLoS Pathog. 9, e1003641 (2013).
Tristem, M., Marshall, C., Karpas, A., Petrik, J. & Hill, F. Origin of vpx in lentiviruses. Nature 347, 341–342 (1990).
Tristem, M., Marshall, C., Karpas, A. & Hill, F. Evolution of the primate lentiviruses: evidence from vpx and vpr. EMBO J. 11, 3405–3412 (1992).
Guyader, M., Emerman, M., Montagnier, L. & Peden, K. VPX mutants of HIV-2 are infectious in established cell lines but display a severe defect in peripheral blood lymphocytes. EMBO J. 8, 1169–1175 (1989).
Yu, X.F., Yu, Q.C., Essex, M. & Lee, T.H. The vpx gene of simian immunodeficiency virus facilitates efficient viral replication in fresh lymphocytes and macrophage. J. Virol. 65, 5088–5091 (1991).
Baldauf, H.M. et al. SAMHD1 restricts HIV-1 infection in resting CD4+ T cells. Nat. Med. 18, 1682–1687 (2012).
Descours, B. et al. SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4+ T-cells. Retrovirology 9, 87 (2012).
Goujon, C. et al. With a little help from a friend: increasing HIV transduction of monocyte-derived dendritic cells with virion-like particles of SIV(MAC). Gene Ther. 13, 991–994 (2006).
Sunseri, N., O'Brien, M., Bhardwaj, N. & Landau, N.R. Human immunodeficiency virus type 1 modified to package Simian immunodeficiency virus Vpx efficiently infects macrophages and dendritic cells. J. Virol. 85, 6263–6274 (2011).
Bobadilla, S., Sunseri, N. & Landau, N.R. Efficient transduction of myeloid cells by an HIV-1-derived lentiviral vector that packages the Vpx accessory protein. Gene Ther. 20, 514–520 (2013).
Durand, S. et al. Tailored HIV-1 vectors for genetic modification of primary human dendritic cells and monocytes. J. Virol. 87, 234–242 (2013).
Norton, T.D., Miller, E.A., Bhardwaj, N. & Landau, N.R. Vpx-containing dendritic cell vaccine induces CTLs and reactivates latent HIV-1 in vitro. Gene Ther. 22, 11–20 (2015).
Le Rouzic, E. et al. HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4–DDB1 ubiquitin ligase. Cell Cycle 6, 182–188 (2007).
Srivastava, S. et al. Lentiviral Vpx accessory factor targets VprBP/DCAF1 substrate adaptor for cullin 4 E3 ubiquitin ligase to enable macrophage infection. PLoS Pathog. 4, e1000059 (2008).
Laguette, N. et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 (2011).
Hrecka, K. et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661 (2011).
Behrendt, R. et al. Mouse SAMHD1 has antiretroviral activity and suppresses a spontaneous cell-intrinsic antiviral response. Cell Reports 4, 689–696 (2013).
Rehwinkel, J. et al. SAMHD1-dependent retroviral control and escape in mice. EMBO J. 32, 2454–2462 (2013).
Hofmann, H. et al. The Vpx lentiviral accessory protein targets SAMHD1 for degradation in the nucleus. J. Virol. 86, 12552–12560 (2012).
Brandariz-Nuñez, A. et al. Role of SAMHD1 nuclear localization in restriction of HIV-1 and SIVmac. Retrovirology 9, 49 (2012).
Kim, B., Nguyen, L.A., Daddacha, W. & Hollenbaugh, J.A. Tight interplay among SAMHD1 protein level, cellular dNTP levels, and HIV-1 proviral DNA synthesis kinetics in human primary monocyte-derived macrophages. J. Biol. Chem. 287, 21570–21574 (2012).
Jáuregui, P., Logue, E.C., Schultz, M.L., Fung, S. & Landau, N.R. Degradation of SAMHD1 by Vpx is independent of uncoating. J. Virol. (11 March 2015).
Goldstone, D.C. et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480, 379–382 (2011).
Powell, R.D., Holland, P.J., Hollis, T. & Perrino, F.W. Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J. Biol. Chem. 286, 43596–43600 (2011).
Lahouassa, H. et al. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 13, 223–228 (2012).
Gramberg, T. et al. Restriction of diverse retroviruses by SAMHD1. Retrovirology 10, 26 (2013).
Pauls, E. et al. Cell cycle control and HIV-1 susceptibility are linked by CDK6-dependent CDK2 phosphorylation of SAMHD1 in myeloid and lymphoid cells. J. Immunol. 193, 1988–1997 (2014).
St Gelais, C. et al. Identification of cellular proteins interacting with the retroviral restriction factor SAMHD1. J. Virol. 88, 5834–5844 (2014).
White, T.E. et al. The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe 13, 441–451 (2013).
Cribier, A., Descours, B., Valadao, A.L., Laguette, N. & Benkirane, M. Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Reports 3, 1036–1043 (2013).
Beloglazova, N. et al. Nuclease activity of the human SAMHD1 protein implicated in the Aicardi-Goutieres syndrome and HIV-1 restriction. J. Biol. Chem. 288, 8101–8110 (2013).
Ryoo, J. et al. The ribonuclease activity of SAMHD1 is required for HIV-1 restriction. Nat. Med. 20, 936–941 (2014).
Hofmann, H. et al. Inhibition of CUL4A Neddylation causes a reversible block to SAMHD1-mediated restriction of HIV-1. J. Virol. 87, 11741–11750 (2013).
Mlcochova, P., Watters, S.A., Towers, G.J., Noursadeghi, M. & Gupta, R.K. Vpx complementation of 'non-macrophage tropic' R5 viruses reveals robust entry of infectious HIV-1 cores into macrophages. Retrovirology 11, 25 (2014).
Crow, Y.J. et al. Characterization of human disease phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1. Am. J. Med. Genet. A. 167, 296–312 (2015).
Stetson, D.B., Ko, J.S., Heidmann, T. & Medzhitov, R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134, 587–598 (2008).
Gao, D. et al. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 341, 903–906 (2013).
Yan, N., Regalado-Magdos, A.D., Stiggelbout, B., Lee-Kirsch, M.A. & Lieberman, J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat. Immunol. 11, 1005–1013 (2010).
Zhao, K. et al. Modulation of LINE-1 and Alu/SVA retrotransposition by Aicardi-Goutieres syndrome-related SAMHD1. Cell Rep. 4, 1108–1115 (2013).
Lim, E.S. et al. The ability of primate lentiviruses to degrade the monocyte restriction factor SAMHD1 preceded the birth of the viral accessory protein Vpx. Cell Host Microbe 11, 194–204 (2012).
Etienne, L., Hahn, B.H., Sharp, P.M., Matsen, F.A. & Emerman, M. Gene loss and adaptation to hominids underlie the ancient origin of HIV-1. Cell Host Microbe 14, 85–92 (2013).
Lenzi, G.M., Domaoal, R.A., Kim, D., Schinazi, R.F. & Kim, B. Kinetic variations between reverse transcriptases of viral protein X coding and noncoding lentiviruses. Retrovirology 11, 111 (2014).
Kyei, G.B., Cheng, X., Ramani, R. & Ratner, L. Cyclin L2 is a critical HIV dependency factor in macrophages that controls SAMHD1 abundance. Cell Host Microbe 17, 98–106 (2015).
Westmoreland, S.V. et al. SIV vpx is essential for macrophage infection but not for development of AIDS. PLoS ONE 9, e84463 (2014).
Strebel, K., Klimkait, T., Maldarelli, F. & Martin, M.A. Molecular and biochemical analyses of human immunodeficiency virus type 1 vpu protein. J. Virol. 63, 3784–3791 (1989).
Terwilliger, E.F., Cohen, E.A., Lu, Y.C., Sodroski, J.G. & Haseltine, W.A. Functional role of human immunodeficiency virus type 1 vpu. Proc. Natl. Acad. Sci. USA 86, 5163–5167 (1989).
Klimkait, T., Strebel, K., Hoggan, M.D., Martin, M.A. & Orenstein, J.M. The human immunodeficiency virus type 1-specific protein vpu is required for efficient virus maturation and release. J. Virol. 64, 621–629 (1990).
Willey, R.L., Maldarelli, F., Martin, M.A. & Strebel, K. Human immunodeficiency virus type 1 Vpu protein regulates the formation of intracellular gp160–CD4 complexes. J. Virol. 66, 226–234 (1992).
Neil, S.J., Eastman, S.W., Jouvenet, N. & Bieniasz, P.D. HIV-1 Vpu promotes release and prevents endocytosis of nascent retrovirus particles from the plasma membrane. PLoS Pathog. 2, e39 (2006).
Sakai, H., Tokunaga, K., Kawamura, M. & Adachi, A. Function of human immunodeficiency virus type 1 Vpu protein in various cell types. J. Gen. Virol. 76, 2717–2722 (1995).
Neil, S.J., Sandrin, V., Sundquist, W.I. & Bieniasz, P.D. An interferon-alpha-induced tethering mechanism inhibits HIV-1 and Ebola virus particle release but is counteracted by the HIV-1 Vpu protein. Cell Host Microbe 2, 193–203 (2007).
Varthakavi, V., Smith, R.M., Bour, S.P., Strebel, K. & Spearman, P. Viral protein U counteracts a human host cell restriction that inhibits HIV-1 particle production. Proc. Natl. Acad. Sci. USA 100, 15154–15159 (2003).
Neil, S.J., Zang, T. & Bieniasz, P.D. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451, 425–430 (2008).
Van Damme, N. et al. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3, 245–252 (2008).
Hinz, A. et al. Structural basis of HIV-1 tethering to membranes by the BST-2/tetherin ectodomain. Cell Host Microbe 7, 314–323 (2010).
Venkatesh, S. & Bieniasz, P.D. Mechanism of HIV-1 virion entrapment by tetherin. PLoS Pathog. 9, e1003483 (2013).
Perez-Caballero, D. et al. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell 139, 499–511 (2009).
Galão, R.P., Le Tortorec, A., Pickering, S., Kueck, T. & Neil, S.J. Innate sensing of HIV-1 assembly by Tetherin induces NFκB-dependent proinflammatory responses. Cell Host Microbe 12, 633–644 (2012).
Cocka, L.J. & Bates, P. Identification of alternatively translated Tetherin isoforms with differing antiviral and signaling activities. PLoS Pathog. 8, e1002931 (2012).
Tokarev, A. et al. Stimulation of NF-κB activity by the HIV restriction factor BST2. J. Virol. 87, 2046–2057 (2013).
Sauter, D. et al. Differential regulation of NF-κB-mediated proviral and antiviral host gene expression by primate lentiviral Nef and Vpu proteins. Cell Reports 10, 586–599 (2015).
Pickering, S. et al. Preservation of tetherin and CD4 counter-activities in circulating Vpu alleles despite extensive sequence variation within HIV-1 infected individuals. PLoS Pathog. 10, e1003895 (2014).
Jia, B. et al. Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLoS Pathog. 5, e1000429 (2009).
Zhang, F. et al. Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe 6, 54–67 (2009).
Lim, E.S., Malik, H.S. & Emerman, M. Ancient adaptive evolution of tetherin shaped the functions of Vpu and Nef in human immunodeficiency virus and primate lentiviruses. J. Virol. 84, 7124–7134 (2010).
Sauter, D. et al. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6, 409–421 (2009).
Kluge, S.F. et al. The transmembrane domain of HIV-1 Vpu is sufficient to confer anti-tetherin activity to SIVcpz and SIVgor Vpu proteins: cytoplasmic determinants of Vpu function. Retrovirology 10, 32 (2013).
Bour, S. & Strebel, K. The human immunodeficiency virus (HIV) type 2 envelope protein is a functional complement to HIV type 1 Vpu that enhances particle release of heterologous retroviruses. J. Virol. 70, 8285–8300 (1996).
Bour, S., Schubert, U., Peden, K. & Strebel, K. The envelope glycoprotein of human immunodeficiency virus type 2 enhances viral particle release: a Vpu-like factor? J. Virol. 70, 820–829 (1996).
Serra-Moreno, R., Jia, B., Breed, M., Alvarez, X. & Evans, D.T. Compensatory changes in the cytoplasmic tail of gp41 confer resistance to tetherin/BST-2 in a pathogenic nef-deleted SIV. Cell Host Microbe 9, 46–57 (2011).
Acknowledgements
We thank M. Ooms and L.C.F. Mulder for critical reading of the manuscript. The work was supported by US National Institutes of Health/National Institute of Allergy and Infectious Disease grants to V.S. (AI064001, AI090935) and N.R.L. (AI067059, AI058864), the Vilcek Fellowship Endowment Fund to N.B. and the American Foundation for AIDS Research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Simon, V., Bloch, N. & Landau, N. Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat Immunol 16, 546–553 (2015). https://doi.org/10.1038/ni.3156
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3156
This article is cited by
-
A functional map of HIV-host interactions in primary human T cells
Nature Communications (2022)
-
Antiviral immunity and nucleic acid sensing in haematopoietic stem cell gene engineering
Gene Therapy (2021)
-
Advancing targeted protein degradation for cancer therapy
Nature Reviews Cancer (2021)
-
Comparative proteomics identifies Schlafen 5 (SLFN5) as a herpes simplex virus restriction factor that suppresses viral transcription
Nature Microbiology (2021)
-
Studying Viral Populations with Tools from Quantum Spin Chains
Journal of Statistical Physics (2021)