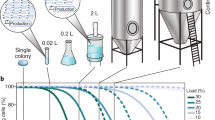
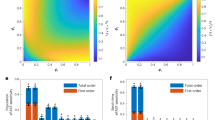
Abstract
Biosynthesis enables renewable production of manifold compounds, yet often biosynthetic performance must be improved for it to be economically feasible. Nongenetic, cell-to-cell variations in protein and metabolite concentrations are naturally inherent, suggesting the existence of both high- and low-performance variants in all cultures. Although having an intrinsic source of low performers might cause suboptimal ensemble biosynthesis, the existence of high performers suggests an avenue for performance enhancement. Here we develop in vivo population quality control (PopQC) to continuously select for high-performing, nongenetic variants. We apply PopQC to two biosynthetic pathways using two alternative design principles and demonstrate threefold enhanced production of both free fatty acid (FFA) and tyrosine. We confirm that PopQC improves ensemble biosynthesis by selecting for nongenetic high performers. Additionally, we use PopQC in fed-batch FFA production and achieve 21.5 g l−1 titer and 0.5 g l−1 h−1 productivity. Given the ubiquity of nongenetic variation, PopQC should be applicable to a variety of metabolic pathways for enhanced biosynthesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Schirmer, A., Rude, M.A., Li, X., Popova, E. & del Cardayre, S.B. Microbial biosynthesis of alkanes. Science 329, 559–562 (2010).
Gronenberg, L.S., Marcheschi, R.J. & Liao, J.C. Next generation biofuel engineering in prokaryotes. Curr. Opin. Chem. Biol. 17, 462–471 (2013).
Woolston, B.M., Edgar, S. & Stephanopoulos, G. Metabolic engineering: past and future. Annu. Rev. Chem. Biomol. Eng. 4, 259–288 (2013).
Paddon, C.J. & Keasling, J.D. Semi-synthetic artemisinin: a model for the use of synthetic biology in pharmaceutical development. Nat. Rev. Microbiol. 12, 355–367 (2014).
Kim, E., Moore, B.S. & Yoon, Y.J. Reinvigorating natural product combinatorial biosynthesis with synthetic biology. Nat. Chem. Biol. 11, 649–659 (2015).
Nielsen, J. et al. Engineering synergy in biotechnology. Nat. Chem. Biol. 10, 319–322 (2014).
Na, D. et al. Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nat. Biotechnol. 31, 170–174 (2013).
Lidstrom, M.E. & Konopka, M.C. The role of physiological heterogeneity in microbial population behavior. Nat. Chem. Biol. 6, 705–712 (2010).
Müller, S., Harms, H. & Bley, T. Origin and analysis of microbial population heterogeneity in bioprocesses. Curr. Opin. Biotechnol. 21, 100–113 (2010).
Taniguchi, Y. et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329, 533–538 (2010).
Li, G.W. & Xie, X.S. Central dogma at the single-molecule level in living cells. Nature 475, 308–315 (2011).
Guimaraes, J.C., Rocha, M. & Arkin, A.P. Transcript level and sequence determinants of protein abundance and noise in Escherichia coli. Nucleic Acids Res. 42, 4791–4799 (2014).
Zenobi, R. Single-cell metabolomics: analytical and biological perspectives. Science 342, 1243259 (2013).
Paige, J.S., Nguyen-Duc, T., Song, W. & Jaffrey, S.R. Fluorescence imaging of cellular metabolites with RNA. Science 335, 1194 (2012).
Love, K.R., Panagiotou, V., Jiang, B., Stadheim, T.A. & Love, J.C. Integrated single-cell analysis shows Pichia pastoris secretes protein stochastically. Biotechnol. Bioeng. 106, 319–325 (2010).
Mustafi, N., Grünberger, A., Kohlheyer, D., Bott, M. & Frunzke, J. The development and application of a single-cell biosensor for the detection of L-methionine and branched-chain amino acids. Metab. Eng. 14, 449–457 (2012).
Labhsetwar, P., Cole, J.A., Roberts, E., Price, N.D. & Luthey-Schulten, Z.A. Heterogeneity in protein expression induces metabolic variability in a modeled Escherichia coli population. Proc. Natl. Acad. Sci. USA 110, 14006–14011 (2013).
Delvigne, F., Zune, Q., Lara, A.R., Al-Soud, W. & Sørensen, S.J. Metabolic variability in bioprocessing: implications of microbial phenotypic heterogeneity. Trends Biotechnol. 32, 608–616 (2014).
Lu, X., Vora, H. & Khosla, C. Overproduction of free fatty acids in E. coli: implications for biodiesel production. Metab. Eng. 10, 333–339 (2008).
Xu, P. et al. Modular optimization of multi-gene pathways for fatty acids production in E. coli. Nat. Commun. 4, 1409 (2013).
Blazeck, J. et al. Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nat. Commun. 5, 3131 (2014).
Zhang, F., Carothers, J.M. & Keasling, J.D. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat. Biotechnol. 30, 354–359 (2012).
Lawrence, M.S., Phillips, K.J. & Liu, D.R. Supercharging proteins can impart unusual resilience. J. Am. Chem. Soc. 129, 10110–10112 (2007).
Lütke-Eversloh, T., Santos, C.N. & Stephanopoulos, G. Perspectives of biotechnological production of L-tyrosine and its applications. Appl. Microbiol. Biotechnol. 77, 751–762 (2007).
Pittard, J., Camakaris, H. & Yang, J. The TyrR regulon. Mol. Microbiol. 55, 16–26 (2005).
Liu, D., Xiao, Y., Evans, B.S. & Zhang, F. Negative feedback regulation of fatty acid production based on a malonyl-CoA sensor-actuator. ACS Synth. Biol. 4, 132–140 (2015).
Doroshenko, V. et al. YddG from Escherichia coli promotes export of aromatic amino acids. FEMS Microbiol. Lett. 275, 312–318 (2007).
Chou, H.H. & Keasling, J.D. Programming adaptive control to evolve increased metabolite production. Nat. Commun. 4, 2595 (2013).
Conrad, T.M. et al. RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media. Proc. Natl. Acad. Sci. USA 107, 20500–20505 (2010).
Nakata, K., Koh, M.M., Tsuchido, T. & Matsumura, Y. All genomic mutations in the antimicrobial surfactant-resistant mutant, Escherichia coli OW66, are involved in cell resistance to surfactant. Appl. Microbiol. Biotechnol. 87, 1895–1905 (2010).
Foster, P.L. Stress-induced mutagenesis in bacteria. Crit. Rev. Biochem. Mol. Biol. 42, 373–397 (2007).
Dietrich, J.A., Shis, D.L., Alikhani, A. & Keasling, J.D. Transcription factor-based screens and synthetic selections for microbial small-molecule biosynthesis. ACS Synth. Biol. 2, 47–58 (2013).
Raman, S., Rogers, J.K., Taylor, N.D. & Church, G.M. Evolution-guided optimization of biosynthetic pathways. Proc. Natl. Acad. Sci. USA 111, 17803–17808 (2014).
Veening, J.W., Smits, W.K. & Kuipers, O.P. Bistability, epigenetics, and bet-hedging in bacteria. Annu. Rev. Microbiol. 62, 193–210 (2008).
Jablonka, E. & Raz, G. Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Q. Rev. Biol. 84, 131–176 (2009).
Kiviet, D.J. et al. Stochasticity of metabolism and growth at the single-cell level. Nature 514, 376–379 (2014).
Keasling, J.D. Manufacturing molecules through metabolic engineering. Science 330, 1355–1358 (2010).
Tanaka, A. & Nakajima, H. Application of immobilized growing cells. Adv. Biochem. Eng. Biotechnol. 42, 97–131 (1990).
Barber, W.P. & Stuckey, D.C. The use of the anaerobic baffled reactor (ABR) for wastewater treatment: a review. Water Res. 33, 1559–1578 (1999).
Dahl, R.H. et al. Engineering dynamic pathway regulation using stress-response promoters. Nat. Biotechnol. 31, 1039–1046 (2013).
Zhang, F. & Keasling, J. Biosensors and their applications in microbial metabolic engineering. Trends Microbiol. 19, 323–329 (2011).
Fernandes, R.L. et al. Experimental methods and modeling techniques for description of cell population heterogeneity. Biotechnol. Adv. 29, 575–599 (2011).
van Heerden, J.H. et al. Lost in transition: start-up of glycolysis yields subpopulations of nongrowing cells. Science 343, 1245114 (2014).
Wang, B.L. et al. Microfluidic high-throughput culturing of single cells for selection based on extracellular metabolite production or consumption. Nat. Biotechnol. 32, 473–478 (2014).
Levine, E. & Hwa, T. Stochastic fluctuations in metabolic pathways. Proc. Natl. Acad. Sci. USA 104, 9224–9229 (2007).
Oyarzún, D.A., Lugagne, J.B. & Stan, G.B. Noise propagation in synthetic gene circuits for metabolic control. ACS Synth. Biol. 4, 116–125 (2015).
Lee, T.S. et al. BglBrick vectors and datasheets: a synthetic biology platform for gene expression. J. Biol. Eng. 5, 12 (2011).
Engler, C., Kandzia, R. & Marillonnet, S. A one pot, one step, precision cloning method with high throughput capability. PLoS One 3, e3647 (2008).
Kempe, K., Hsu, F.F., Bohrer, A. & Turk, J. Isotope dilution mass spectrometric measurements indicate that arachidonylethanolamide, the proposed endogenous ligand of the cannabinoid receptor, accumulates in rat brain tissue post mortem but is contained at low levels in or is absent from fresh tissue. J. Biol. Chem. 271, 17287–17295 (1996).
Juminaga, D. et al. Modular engineering of L-tyrosine production in Escherichia coli. Appl. Environ. Microbiol. 78, 89–98 (2012).
Acknowledgements
The authors acknowledge D. Liu at Harvard University for the GFP reporter gene. The authors would like to thank K. Naegle, C. Immethun, A. Hoynes-O'Connor, D. Giblin, F.-F. Hsu and E. Lantelme for technical assistance and the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for helping with genomic analysis. This work was supported by a start-up package from Washington University, the Defense Advanced Research Projects Agency (D13AP00038 to F.Z.), the National Science Foundation (MCB1453147 and MCB1331194, both to F.Z.), the Human Frontier Science Program (RGY0076/2015 to F.Z.) and the International Center for Advanced Renewable Energy and Sustainability (I-CARES).
Author information
Authors and Affiliations
Contributions
F.Z. conceived the project. F.Z., Y.X. and C.H.B. designed the experiments. Y.X. and C.H.B. performed the experiments. F.Z., Y.X., C.H.B. and D.L. analyzed the data and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
Y.X. and F.Z. have filed a patent application ("A Genetically-Encoded Quality Control System for Improving the Microbial Production of Chemicals, Pharmaceuticals and Fuels," US Provisional Patent Application Ser. No. #62/214,248) on the basis of this contribution.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–12, Supplementary Tables 1–4 and Supplementary Note. (PDF 2137 kb)
Rights and permissions
About this article
Cite this article
Xiao, Y., Bowen, C., Liu, D. et al. Exploiting nongenetic cell-to-cell variation for enhanced biosynthesis. Nat Chem Biol 12, 339–344 (2016). https://doi.org/10.1038/nchembio.2046
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2046
This article is cited by
-
Method for plasmid-based antibiotic-free fermentation
Microbial Cell Factories (2024)
-
Heterologous production of 3-hydroxypropionic acid in Methylorubrum extorquens by introducing the mcr gene via a multi-round chromosomal integration system based on cre-lox71/lox66 and transposon
Microbial Cell Factories (2024)
-
Metabolic stress constrains microbial L-cysteine production in Escherichia coli by accelerating transposition through mobile genetic elements
Microbial Cell Factories (2023)
-
Microbial engineering strategies to utilize waste feedstock for sustainable bioproduction
Nature Reviews Bioengineering (2023)
-
Strain and process engineering toward continuous industrial fermentation
Frontiers of Chemical Science and Engineering (2023)