Abstract
The auto-phosphorylation of murine receptor-interacting protein 3 (Rip3) on Thr 231 and Ser 232 in the necrosome is required to trigger necroptosis. However, how Rip3 phosphorylation is regulated is still largely unknown. Here we identified protein phosphatase 1B (Ppm1b) as a Rip3 phosphatase and found that Ppm1b restricts necroptosis in two settings: spontaneous necroptosis caused by Rip3 auto-phosphorylation in resting cells, and tumour necrosis factor-α (TNF)-induced necroptosis in cultured cells. We revealed that Ppm1b selectively suppresses necroptosis through the dephosphorylation of Rip3, which then prevents the recruitment of mixed lineage kinase domain-like protein (Mlkl) to the necrosome. We further showed that Ppm1b deficiency (Ppm1bd/d) in mice enhanced TNF-induced death in a Rip3-dependent manner, and the role of Ppm1b in inhibiting necroptosis was evidenced by elevated Rip3 phosphorylation and tissue damage in the caecum of TNF-treated Ppm1bd/d mice. These data indicate that Ppm1b negatively regulates necroptosis through dephosphorylating Rip3 in vitro and in vivo.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Change history
19 March 2015
In the version of this Article originally published online the molecular mass of Ppm1b-S (isoform 2) should have read Mr43K. This has been corrected in all versions of the Article.
References
Vandenabeele, P., Galluzzi, L., Vanden Berghe, T. & Kroemer, G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat. Rev. Mol. Cell. Biol. 11, 700–714 (2010).
Vanlangenakker, N., Bertrand, M. J., Bogaert, P., Vandenabeele, P. & Vanden Berghe, T. TNF-induced necroptosis in L929 cells is tightly regulated by multiple TNFR1 complex I and II members. Cell Death Dis. 2, e230 (2011).
Kaiser, W. et al. Toll-like receptor 3-mediated necrosis via TRIF, RIP3, and MLKL. J. Biol. Chem. 288, 31268–31279 (2013).
Upton, J. W., Kaiser, W. J. & Mocarski, E. S. Virus inhibition of RIP3-dependent necrosis. Cell Host Microbe 7, 302–313 (2010).
Kaczmarek, A., Vandenabeele, P. & Krysko, D. V. Necroptosis: the release of damage-associated molecular patterns and its physiological relevance. Immunity 38, 209–223 (2013).
Galluzzi, L., Kepp, O., Krautwald, S., Kroemer, G. & Linkermann, A. Molecular mechanisms of regulated necrosis. Sem. Cell Dev. Biol. 35, 24–32 (2014).
Holler, N. et al. Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nat. Immunol. 1, 489–495 (2000).
Zhang, D. W. et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science 325, 332–336 (2009).
He, S. et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-α. Cell 137, 1100–1111 (2009).
Cho, Y. S. et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 137, 1112–1123 (2009).
Li, J. et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell 150, 339–350 (2012).
Zhao, J. et al. Mixed lineage kinase domain-like is a key receptor interacting protein 3 downstream component of TNF-induced necrosis. Proc. Natl Acad. Sci. USA 109, 5322–5327 (2012).
Sun, L. et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 148, 213–227 (2012).
Murphy, J. M. et al. The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity 39, 443–453 (2013).
Wang, H. et al. Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP3. Mol. Cell 54, 133–146 (2014).
Dondelinger, Y. et al. MLKL compromises plasma membrane integrity by binding to phosphatidylinositol phosphates. Cell Rep. 7, 971–981 (2014).
Chen, X. et al. Translocation of mixed lineage kinase domain-like protein to plasma membrane leads to necrotic cell death. Cell Res. 24, 105–121 (2014).
Cai, Z. et al. Plasma membrane translocation of trimerized MLKL protein is required for TNF-induced necroptosis. Nat. Cell Biol. 16, 55–65 (2014).
Chen, W. et al. Diverse sequence determinants control human and mouse receptor interacting protein 3 (RIP3) and mixed lineage kinase domain-like (MLKL) interaction in necroptotic signaling. J. Biol. Chem. 288, 16247–16261 (2013).
Wu, C. et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 10, R130 (2009).
Lattin, J. E. et al. Expression analysis of G protein-coupled receptors in mouse macrophages. Immunome Res. 4, 5 (2008).
Terasawa, T. et al. Molecular cloning of a novel isotype of Mg(2+)-dependent protein phosphatase β (type 2C β) enriched in brain and heart. Arch. Biochem. Biophys. 307, 342–349 (1993).
Kusuda, K. et al. Mutational analysis of the domain structure of mouse protein phosphatase 2Cβ. Biochem. J. 332, 243–250 (1998).
Wu, T. et al. Regulator of G-protein signaling 19 (RGS19) and its partner Gα-inhibiting activity polypeptide 3 (GNAI3) are required for zVAD-induced autophagy and cell death in L929 cells. PLoS ONE 9, e94634 (2014).
Degterev, A. et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat. Chem. Biol. 4, 313–321 (2008).
Sun, X., Yin, J., Starovasnik, M. A., Fairbrother, W. J. & Dixit, V. M. Identification of a novel homotypic interaction motif required for the phosphorylation of receptor-interacting protein (RIP) by RIP3. J. Biol. Chem. 277, 9505–9511 (2002).
Chen, J., Zheng, X. F., Brown, E. J. & Schreiber, S. L. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc. Natl Acad. Sci. USA 92, 4947–4951 (1995).
Wu, J. et al. Mlkl knockout mice demonstrate the indispensable role of Mlkl in necroptosis. Cell Res. 23, 994–1006 (2013).
Wu, Y. T. et al. zVAD-induced necroptosis in L929 cells depends on autocrine production of TNFα mediated by the PKC-MAPKs-AP-1 pathway. Cell Death Differ. 18, 26–37 (2011).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Sasaki, M. et al. Disruption of the mouse protein Ser/Thr phosphatase 2Cβ gene leads to early pre-implantation lethality. Mech. Dev. 124, 489–499 (2007).
Newton, K. et al. Activity of protein kinase RIPK3 determines whether cells die by necroptosis or apoptosis. Science 343, 1357–1360 (2014).
Wang, L., Du, F. & Wang, X. TNF-α induces two distinct caspase-8 activation pathways. Cell 133, 693–703 (2008).
Bertrand, M. J. et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol. Cell 30, 689–700 (2008).
Sun, W. et al. PPM1A and PPM1B act as IKKβ phosphatases to terminate TNFα-induced IKKβ-NF-κB activation. Cell Signal. 21, 95–102 (2009).
Podolin, P. L. et al. Attenuation of murine collagen-induced arthritis by a novel, potent, selective small molecule inhibitor of IκB Kinase 2, TPCA-1 (2-[(aminocarbonyl)amino]-5-(4-fluorophenyl)-3-thiophenecarboxamide), occurs via reduction of proinflammatory cytokines and antigen-induced T cell Proliferation. J. Pharmacol. Exp. Ther. 312, 373–381 (2005).
Tanaka, A. et al. A novel NF-κB inhibitor, IMD-0354, suppresses neoplastic proliferation of human mast cells with constitutively activated c-kit receptors. Blood 105, 2324–2331 (2005).
Hanada, M. et al. Regulation of the TAK1 signaling pathway by protein phosphatase 2C. J. Biol. Chem. 276, 5753–5759 (2001).
Ninomiya-Tsuji, J. et al. A resorcylic acid lactone, 5Z-7-oxozeaenol, prevents inflammation by inhibiting the catalytic activity of TAK1 MAPK kinase kinase. J. Biol. Chem. 278, 18485–18490 (2003).
Xie, T. et al. Structural insights into RIP3-mediated necroptotic signaling. Cell Rep. 5, 70–78 (2013).
Li, L. et al. The Gβγ-Src signaling pathway regulates TNF-induced necroptosis via control of necrosome translocation. Cell Res. 24, 417–432 (2014).
Lin, X. et al. PPM1A functions as a Smad phosphatase to terminate TGFβ signaling. Cell 125, 915–928 (2006).
Linkermann, A. et al. Dichotomy between RIP1- and RIP3-mediated necroptosis in tumor necrosis factor-α-induced shock. Mol. Med. 18, 577–586 (2012).
Duprez, L. et al. RIP kinase-dependent necrosis drives lethal systemic inflammatory response syndrome. Immunity 35, 908–918 (2011).
Tracey, K. J. et al. Shock and tissue injury induced by recombinant human cachectin. Science 234, 470–474 (1986).
Choi, H. K. et al. PKA negatively regulates PP2Cβ to activate NF-κB-mediated inflammatory signaling. Biochem. Biophys. Res. Commun. 436, 473–477 (2013).
Klumpp, S., Selke, D. & Hermesmeier, J. Protein phosphatase type 2C active at physiological Mg2+: stimulation by unsaturated fatty acids. FEBS Lett. 437, 229–232 (1998).
Shreeram, S. et al. Wip1 phosphatase modulates ATM-dependent signaling pathways. Mol. Cell 23, 757–764 (2006).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Zhang, X., Goncalves, R. & Mosser, D. M. The isolation and characterization of murine macrophages. Curr. Protoc. Immunol. 11 (2008).
Wu, X. et al. Investigation of RIP3-dependent protein phosphorylation by quantitative phosphoproteomics. Mol. Cell Proteomics 11, 1640–1651 (2012).
Coburn, B., Li, Y., Owen, D., Vallance, B. A. & Finlay, B. B. Salmonella enterica serovar Typhimurium pathogenicity island 2 is necessary for complete virulence in a mouse model of infectious enterocolitis. Infect. Immun. 73, 3219–3227 (2005).
Acknowledgements
We thank S. Tamura and M. Ohnishi for Ppm1b+/− mice. This work was supported by the National Basic Research Program of China (973 Program; 2015CB553800), the National Scientific and Technological Major Project (2013ZX10002-002), the National Natural Science Foundation of China (91429301, 31420103910, 31330047, 91029304, 31221065, and 31090360), the Hi-Tech Research and Development Program of China (863 program; 2012AA02A201), the 111 Project (B12001), the Science and Technology Foundation of Xiamen (No. 3502Z20130027), the National Science Foundation of China for Fostering Talents in Basic Research (Grant No.J1310027) and the Open Research Fund of State Key Laboratory of Cellular Stress Biology, Xiamen University. This research was also partly supported by grants from MOST (2012CB966600) and NIH (R01GM63773, R01AR053591, R01CA108454). The collaborative research is also supported by the special funds for Innovation Center for Cell Biology from Zhejiang University and Xiamen University.
Author information
Authors and Affiliations
Contributions
W.C., J.W., L.L., Zhengmo Z., J.R., Y.L., X-H.F. and J.H. carried out the experiments. F.C., C.Y., Zhenru Z., S.S.S., X.Z., Zhirong Z., C-Q.Z., H.W., M.X. and X.L. helped to prepare cell lines, generated the ppm1b gene-trap line, provided reagents and mass spectrum analysis. J.H. contributed to the overall design of the project. W.C. and J.H. interpreted the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
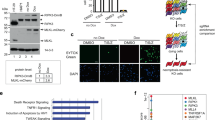
Supplementary Figure 1 Ppm1b interacts with Rip3.
(a) Ppm1b interacts with Rip3 in resting stage in L929 cells. Rip3-KO–Flag–Rip3 L929 cells were lysed and the cell lysates were immunoprecipitated with anti-HA (IP: HA) and anti-Flag (IP: Flag) antibodies. Both the cell lysate (Input) and immunoprecipitates were analysed by immunoblotting with indicated antibodies. The vertical line represents a splice mark. The samples were obtained and processed in the same experiment, and the gels/blots were processed in parallel. (b) Both Ppm1b-L and Ppm1b-S co-immunoprecipitate with Rip3 in 293T cells. Myc-Rip3 was coexpressed with Flag-tagged Ppm1b-L, Ppm1b-S or empty vector. Then co-immunoprecipitation experiments were performed with anti-Flag antibody. Both the immunoprecipitates and lysates were analysed by immunoblotting with indicated antibodies. (c) Ppm1b selectively interacts with Rip3 but not the other necrosome components in 293T cells. Flag–Ppm1b-L was coexpressed with HA-tagged proteins as indicated. Co-immunoprecipitaton experiments were performed with anti-HA antibody and analysed as in (b). (d) Schematic of the domain structures of Ppm1b-L and Ppm1b-S and Rip3. (e) The phosphatase domain of Ppm1b interacts with Rip3. Myc-Rip3 was coexpressed with Flag-tagged full length and truncated Ppm1b in 293T cells. Then co-immunoprecipitation experiments were performed as in (b). Ppm1b-L-PD: Ppm1b-L phosphatase domain; Ppm1b-L-CD: Ppm1b-L C-terminal domain. (f) The kinase domain of Rip3 interacts with Ppm1b. Flag–Ppm1b-L-PD was coexpressed with Myc-tagged full length and truncated Rip3 in 293T cells. Then co-immunoprecipitation experiments were performed with anti-Myc antibody and analysed as in (b). Rip3-KD: Rip3 kinase domain; Rip3-CD: Rip3 C-terminal domain. Data shown were representative of two or more independent experiments. Uncropped images of blots are shown in Supplementary Fig. 7.
Supplementary Figure 2 Related to Fig. 2.
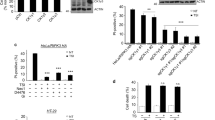
(a) The protein amounts in Ppm1b knocked down and control L929 cells were analysed by immunoblotting with indicated antibodies. Related to Fig. 2a. (b) Ppm1b-S, Ppm1b-L and both isoforms were knocked down with isoform-specific shRNAs in L929 cells. Left, 48 h later, spontaneous cell death was measured. Right, the protein amounts were analysed by immunoblotting with indicated antibodies. Results shown were mean ± s.e.m.;n = 3,000 cells pooled from 3 independent experiments. (c) The protein amounts in Ppm1b knocked down and control cells were analysed by immunoblotting with indicated antibodies. Related to Fig. 2c. (d) The protein amounts in WT and Rip3 KO L929 cells knocked down with shPpm1b or control shRNAs were analysed by immunoblotting with indicated antibodies. Related to Fig. 2f. (e) The protein amounts in WT and Rip3 KO mouse peritoneal macrophage cells knocked down with shPpm1b or control shRNAs were analysed by immunoblotting with indicated antibodies. Related to Fig. 2g. For Figure a, c, d, e, data shown were representative of two or more independent experiments. The asterisk (∗) denotes a nonspecific band. Statistics source data for this figure can be found in Supplementary Table 2. Uncropped images of blots are shown in Supplementary Fig. 7.
Supplementary Figure 3 Related to Fig. 3.
(a) Ppm1b was knocked down in Rip3 KO L929 cells reconstituted with Rip3-WT, Rip3 phosphorylation-deficient mutant (Rip3-2A) mutant and Rip3 RHIM domain mutant (Rip3RHIM). The cells were lysed and subjected to immunoblotting with indicated antibodies. Related to Fig. 3b. (b) Ppm1b was knocked down in WT and Rip1KO L929 cells. The cells were lysed and subjected to immunoblotting with indicated antibodies. The vertical line represents a splice mark. The spliced images were from the same blot. Related to Fig. 3c. (c) WT Rip3 and Rip3RHIM were introduced into Rip1 KO and WT L929 cells by lentiviral vectors. The cells were lysed and subjected to immunoblotting with indicated antibodies. Related to Fig. 3d. (d) Ppm1b was knocked down in WT and MlklKO L929 cells. The cells were lysed and subjected to immunoblotting with indicated antibodies. Related to Fig. 3h. (e) TNFR1 KO and WT L929 cells were infected with lentivirus encoding shPpm1b or control. Left, spontaneous cell death was analysed 48 h later. n = 3,000 cells pooled from three independent experiments. Right, TNFR1 KO and WT L929 cells were treated with or without TNF for 12 h. Cell death was analysed. n = 3 independent experiments. Results shown were mean ± s.e.m.;##P < 0.01;###P < 0.001. For Figure a-d, data shown were representative of two or more independent experiments. The asterisk (∗) denotes a nonspecific band. Statistics source data for this figure can be found in Supplementary Table 2. Uncropped images of blots are shown in Supplementary Fig. 7.
Supplementary Figure 4 Ppm1b targets TNF-induced necroptosis but not apoptosis.
(a,b) L929 cells were infected with lentivirus encoding shPpm1b or control, or not infected (Mock). 48 h later, (a) the cells were treated with TNF (10 ng ml−1) for indicated time periods and cell death was analysed by flow cytometer; (b) the cells were treated with different doses of TNF as indicated for 6 h and cell death was analysed. n = 3 independent experiments. (c) Ppm1b does not affect TNF-induced apoptosis in NIH3T3-A cells. Ppm1b was knocked down in NIH3T3-A cells. Left, 48 h later, the cells were treated with or without TNF (100 ng ml−1) for 24 h and cell death was analysed. n = 3 independent experiments. Right, the cell lysates were analysed by immunoblotting with indicated antibodies. (d) Both Ppm1b-L and S isoforms restrict TNF-induced necroptosis in L929. Ppm1b-S, Ppm1b-L or both isoforms were knocked down with isoform specific shRNAs in L929 cells. 48 h later, the cells were treated with or without TNF (10 ng ml−1) for 6 h and cell death was measured. n = 3 independent experiments. (e) Ppm1b KO L929 cells were infected with different doses of lentivirus encoding Ppm1b-L and Ppm1b-S as indicated, or not infected (Mock infection). Then the cells were treated with TNF (10 ng ml−1) for 6 h and cell death was analysed. n = 3 independent experiments. (f) Ppm1b does not affect the TNF-induced p65 and p38 phosphorylation. Ppm1b was knocked down with shRNA in L929 cells. 48 h later, the cells were treated with TNF (10 ng ml−1) for indicated time. The cell lysates were analysed by immunoblotting with indicated antibodies. (g) The Ppm1b-β-geo fusion mRNA sequences in Ppm1bd/d mice were determined by 3’ RACE. For Figure a-e, results shown were mean ± s.e.m.;#P < 0.05;##P < 0.01;###P < 0.001; NS: no significant difference. For Figure f, data shown were representative of two independent experiments. The asterisk (∗) denotes a nonspecific band. Statistics source data for this figure can be found in Supplementary Table 2. Uncropped images of blots are shown in Supplementary Fig. 7.
Supplementary Figure 5 The regulation of necroptosis by Ppm1b is independent of NF-κB pathway.
(a) HeLa cells were infected with lentivirus encoding sh-hPpm1b or control. 48 h later, the cells were treated with hTNF (30 ng ml−1) for indicated time and subjected to immunoblotting with indicated antibodies. (b) HT29 cells were analysed as in (a). (c) L929 cells were infected with lentivirus encoding shPpm1b or control, or not infected (Mock). 48 h later, the cells were treated with TNF for indicated time and the IL-6 level of the cell culture supernatant was analysed by ELISA. n = 3 independent experiments. (d) Left, L929 cells were infected with lentivirus encoding shPpm1b or control, or not infected (Mock). 5 h after infection, the cells were treated with DMSO, TPCA-1 (1μM) or IMD 0354 (5μM). 48 h later, spontaneous cell death was analysed. Middle, IKKβ KO and WT L929 cells were infected with lentivirus encoding shPpm1b or control, or not infected. 48 h later, spontaneous cell death was analysed. Right, the IKKβ protein amount in IKKβ KO and WT L929 cells was determined by immunoblotting. n = 3,000 cells pooled from 3 independent experiments. (e) Left, the cells were treated as in (d, left), except that the cells were treated with TNF for 5 h and the TNF-induced cell death rather than spontaneous cell death was analysed 48 h after infection. Right, the cells were treated as in (d, middle), except that the cells were treated with TNF for 5 h and TNF-induced cell death rather than spontaneous cell death was analysed 48 h after infection. n = 3 independent experiments. (f) L929 cells were pretreated with DMSO, TPCA-1 or IMD 0354 for 2 h followed by treatment with TNF for indicated time. The IL-6 level of the cell culture supernatant was analysed by ELISA. n = 3 independent experiments. (g) L929 cells were infected with lentivirus encoding shPpm1b or control, or not infected. 48 h later, the cells were pretreated with DMSO or 5z-7 (1 μm) for 2 h followed by treatment with TNF for 5 h. Then the cell death was analysed. n = 3 independent experiments. (h) HeLa cells stably expressing human Rip3 (HeLa-hRip3) were analysed as in (a). (i) HeLa-hRip3 cells were infected with lentivirus encoding sh-hPpm1b or control, or not infected. Spontaneous cell death and hTNF (30 ng ml−1) + Smac mimetic (100 nM) + zVAD (20 μM) (TSZ)-induced cell death were analysed as in (d, left) and (e, left). n = 3 independent experiments. (j) Littermates of Ppm1bd/d and WT mice were injected with TNF (15 μg) via the tail vein. Then the serum IL-6 level was analysed by ELISA at different time points as indicated. n = 4 mice for each group in single experiment where two independent experiments were performed to assess reproducibility. ND: not detectable. For Figure c-g, i, j, results shown were mean ± s.e.m.;#P < 0.05;##P < 0.01;###P < 0.001;. For Figure a, b, h, data shown were representative of two or more independent experiments. Statistics source data for this figure can be found in Supplementary Table 2. Uncropped images of blots are shown in Supplementary Fig. 7.
Supplementary Figure 6 TNF induces tissue damage in different organs of WT and Rip3−/− mice.
Littermates of WT and Rip3−/− mice were injected with TNF (15 μg) via the tail vein for indicated time. The sections of kidney were analysed by PAS staining while those of the other organs were analysed by H&E staining. The representative images were shown (n = 5 mice of each genotype at 12 h time point; n = 3 mice of each genotype at 0 h time point.). Scale bar, 50 or 100 μm as indicated.
Supplementary information
Supplementary Information
Supplementary Information (PDF 2544 kb)
Supplementary Table 1
Supplementary Information (XLSX 37 kb)
Supplementary Table 2
Supplementary Information (XLSX 25 kb)
Rights and permissions
About this article
Cite this article
Chen, W., Wu, J., Li, L. et al. Ppm1b negatively regulates necroptosis through dephosphorylating Rip3. Nat Cell Biol 17, 434–444 (2015). https://doi.org/10.1038/ncb3120
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3120
This article is cited by
-
Roles of RIPK3 in necroptosis, cell signaling, and disease
Experimental & Molecular Medicine (2022)
-
Ketamine inhibits TNF-α-induced cecal damage by enhancing RIP1 ubiquitination to attenuate lethal SIRS
Cell Death Discovery (2022)
-
Salt-inducible kinases inhibitor HG-9-91-01 targets RIPK3 kinase activity to alleviate necroptosis-mediated inflammatory injury
Cell Death & Disease (2022)
-
Deficiency of PPP6C protects TNF-induced necroptosis through activation of TAK1
Cell Death & Disease (2022)
-
Mosaic composition of RIP1–RIP3 signalling hub and its role in regulating cell death
Nature Cell Biology (2022)