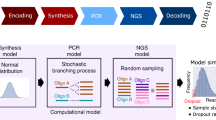
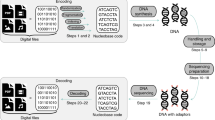
Abstract
Synthetic DNA is durable and can encode digital data with high density, making it an attractive medium for data storage. However, recovering stored data on a large-scale currently requires all the DNA in a pool to be sequenced, even if only a subset of the information needs to be extracted. Here, we encode and store 35 distinct files (over 200 MB of data), in more than 13 million DNA oligonucleotides, and show that we can recover each file individually and with no errors, using a random access approach. We design and validate a large library of primers that enable individual recovery of all files stored within the DNA. We also develop an algorithm that greatly reduces the sequencing read coverage required for error-free decoding by maximizing information from all sequence reads. These advances demonstrate a viable, large-scale system for DNA data storage and retrieval.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
06 March 2018
In the version of this article initially published, the references in the reference list were in the wrong order; the references have been renumbered as follows: 3 as 2; 5 as 3; 6 as 8; 7 as 9; 8 as 11; 9 as 6; 10 as 12; 11 as 5; 12 as 13; 13 as 7; 16 as 10; and no. 2, “Hoch, J.A. & Losick, R. Panspermia, spores and the Bacillus subtilis genome. Nature 390, 237–238 (1997),” has been deleted. In addition, on p.242, end of paragraph 2, the citation in “experiments7” has been deleted. The errors have been corrected in the HTML and PDF versions of the article.
References
Neiman, M.S. On the molecular memory systems and the directed mutations. Radiotekhnika 6, 1–8 (1965).
Cox, J.P.L. Long-term data storage in DNA. Trends Biotechnol. 19, 247–250 (2001).
Church, G.M., Gao, Y. & Kosuri, S. Next-generation digital information storage in DNA. Science 337, 1628 (2012).
Goldman, N. et al. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA. Nature 494, 77–80 (2013).
Grass, R.N., Heckel, R., Puddu, M., Paunescu, D. & Stark, W.J. Robust chemical preservation of digital information on DNA in silica with error-correcting codes. Angew. Chem. Int. Ed. 54, 2552–2555 (2015).
Blawat, M. et al. Forward error correction for DNA data storage. Procedia Comput. Sci. 80, 1011–1022 (2016).
Erlich, Y. & Zielinski, D. DNA Fountain enables a robust and efficient storage architecture. Science 355, 950–954 (2017).
Yazdi, S.M.H.T., Yuan, Y., Ma, J., Zhao, H. & Milenkovic, O. A rewritable, random-access DNA-based storage system. Sci. Rep. 5, 14138 (2015).
Bornholt, J. et al. in Proc. Int. Conf. ASPLOS. 637–649 (ACM, 2016).
Yazdi, S.M.H.T., Gabrys, R. & Milenkovic, O. Portable and error-free DNA-based data storage. Sci. Rep. 7, 5011 (2017).
Kosuri, S. & Church, G.M. Large-scale de novo DNA synthesis: technologies and applications. Nat. Methods 11, 499–507 (2014).
Xu, Q., Schlabach, M.R., Hannon, G.J. & Elledge, S.J. Design of 240,000 orthogonal 25mer DNA barcode probes. Proc. Natl. Acad. Sci. USA 106, 2289–2294 (2009).
Batu, T., Kannan, S., Khanna, S. & McGregor, A. Reconstructing strings from random traces. Proc. Fifteenth Annu. ACM-SIAM SODA'04. 2004, 910–918 (2004).
Pellicer, J., Fay, M.F. & Leitch, I.J. The largest eukaryotic genome of them all? Bot. J. Linn. Soc. 164, 10–15 (2010).
Zadeh, J.N. et al. NUPACK: analysis and design of nucleic acid systems. J. Comput. Chem. 32, 170–173 (2011).
Acknowledgements
We would like to thank B. Peck, P. Finn, S. Chen, A. Stewart, B. Arias, and E. Leproust from Twist Bioscience for supplying the DNA, suggesting protocol refinements, and offering input to our data analysis. We also thank J. Bornholt, K. D'Silva, and A. Levskaya for their help in the early stages of this project, and Y. Chou for her help in preparing samples for distribution. This work was supported in part by a sponsored research agreement by Microsoft, NSF award CCF-1409831 to L.C. and G.S. and by NSF award CCF-1317653 to G.S.
Author information
Authors and Affiliations
Contributions
L.O., Y.J.C., and R.L. designed protocols and performed experiments. S.Y., S.D.A., K.M., M.Z.R., C.R., and P.G. designed and implemented the encoding and decoding pipeline. S.D.A., M.Z.R., G.K., Ke.S., and C.N.T., collected and analyzed data. B.N., C.N.T., S.N., G.G., H.Y.P., R.C., and J.M. assisted in designing and evaluating experiments. D.C., G.S., L.C., and Ka.S. designed experiments, analyzed data and supervised the work.
Corresponding authors
Ethics declarations
Competing interests
S.D.A., Y.-J.C., S.Y., K.M., M.Z.R., G.K., P.G., B.N., H.-Y.P., C.R., G.G., R.C., J.M., D.C., and K.S. are or were employees at Microsoft Research.
Integrated supplementary information
Supplementary Figure 1 Primer sequence design.
(a) Method for primer design. A random 20-mer continues to mutate until it satisfies the design criteria explained above. After satisfying these criteria, the primer is filtered by secondary structure and melting temperature. After generating a library of primers, the library is screened using BLAST to further improve sequence orthogonality. (b) Example of scoring for a primer. If the primer violates a design criterion, all bases related to the violation receive a +1 score.
Supplementary Figure 2 Primer library scalability estimates.
(a) The total number of primer pairs that pass the selection criteria described in Supplementary Fig. 1 (y-axis) increases with the log of the number of starting random 20-mers (x-axis, logarithmic scale). The six blue dots are primer libraries generated from different numbers of starting random 20-mers. (b) The ratio of primers passing the primer-payload collision detection algorithm described in Supplementary Fig. 4 (y-axis) decreases as the log of the amount of information to be stored increases (x-axis, logarithmic scale). Blue points represent the average passing ratio of the six different primer libraries generated in a. Error bars indicate standard deviation calculated from these six primer libraries. The measures of the centre indicate mean calculated from these six primer libraries.
Supplementary Figure 3 Randomization algorithm.
Digital data are iteratively randomized to reduce collisions between primers and payloads.
Supplementary Figure 4 Comparing amplification of files with and without collision detection and our primer design method.
All 15 bp traces are size markers used by the Qiagen QIAxcel system. The same instrument and profile was used for each trace. Each trace is a representative of three independent trials with virtually identical results. (a) “Simple conditions” indicates single-file pools. i. Trace of an amplified file designed with collision detection. ii. Trace of an amplified file designed without collision detection. (b) “Complex conditions” indicates multi-file pools. i. Trace of an amplified file designed with collision detection (9-file pool; amplified file is 17.4% of the pool). ii. Trace of an amplified file designed without using collision detection (6-file pool; amplified file is 18.0% of the pool).
Supplementary Figure 5 Random access and library preparation layout for sequencing.
First, random access regions are used to select files for sequencing. Through ePCR, a 25N region is added to the oligos to improve nucleotide diversity. Then, samples are ligated to Illumina sequencing adaptors with modified Illumina TruSeq Nano kit protocol. Finally, prepared samples are sequenced on an Illumina NextSeq instrument with a 10%-20% PhiX spike-in.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 (PDF 565 kb)
Rights and permissions
About this article
Cite this article
Organick, L., Ang, S., Chen, YJ. et al. Random access in large-scale DNA data storage. Nat Biotechnol 36, 242–248 (2018). https://doi.org/10.1038/nbt.4079
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.4079
This article is cited by
-
DNA as a universal chemical substrate for computing and data storage
Nature Reviews Chemistry (2024)
-
Reconstruction algorithms for DNA-storage systems
Scientific Reports (2024)
-
Recent Progress in High-Throughput Enzymatic DNA Synthesis for Data Storage
BioChip Journal (2024)
-
Modelling for Efficient Scientific Data Storage Using Simple Graphs in DNA
SN Computer Science (2024)
-
In-vitro validated methods for encoding digital data in deoxyribonucleic acid (DNA)
BMC Bioinformatics (2023)