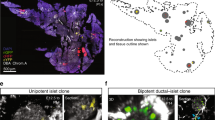
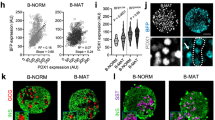
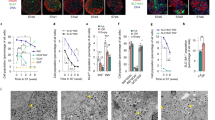
Abstract
Insulin-dependent diabetes is a complex multifactorial disorder characterized by loss or dysfunction of β-cells. Pancreatic β-cells differ in size, glucose responsiveness, insulin secretion and precursor cell potential1,2,3,4,5; understanding the mechanisms that underlie this functional heterogeneity might make it possible to develop new regenerative approaches. Here we show that Fltp (also known as Flattop and Cfap126), a Wnt/planar cell polarity (PCP) effector and reporter gene6, acts as a marker gene that subdivides endocrine cells into two subpopulations and distinguishes proliferation-competent from mature β-cells with distinct molecular, physiological and ultrastructural features. Genetic lineage tracing revealed that endocrine subpopulations from Fltp-negative and -positive lineages react differently to physiological and pathological changes. The expression of Fltp increases when endocrine cells cluster together to form polarized and mature 3D islet mini-organs7,8,9. We show that 3D architecture and Wnt/PCP ligands are sufficient to trigger β-cell maturation. By contrast, the Wnt/PCP effector Fltp is not necessary for β-cell development, proliferation or maturation. We conclude that 3D architecture and Wnt/PCP signalling underlie functional β-cell heterogeneity and induce β-cell maturation. The identification of Fltp as a marker for endocrine subpopulations sheds light on the molecular underpinnings of islet cell heterogeneity and plasticity and might enable targeting of endocrine subpopulations for the regeneration of functional β-cell mass in diabetic patients.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Salomon, D. & Meda, P. Heterogeneity and contact-dependent regulation of hormone secretion by individual B cells. Exp. Cell Res. 162, 507–520 (1986)
Heimberg, H. et al. Heterogeneity in glucose sensitivity among pancreatic β-cells is correlated to differences in glucose phosphorylation rather than glucose transport. EMBO J. 12, 2873–2879 (1993)
Smukler, S. R. et al. The adult mouse and human pancreas contain rare multipotent stem cells that express insulin. Cell Stem Cell 8, 281–293 (2011)
Razavi, R. et al. Diabetes enhances the proliferation of adult pancreatic multipotent progenitor cells and biases their differentiation to more β-cell production. Diabetes 64, 1311–1323 (2015)
Katsuta, H. et al. Single pancreatic β cells co-express multiple islet hormone genes in mice. Diabetologia 53, 128–138 (2010)
Gegg, M. et al. Flattop regulates basal body docking and positioning in mono- and multiciliated cells. eLife 3, e03842 (2014)
Bonner-Weir, S. Morphological evidence for pancreatic polarity of β-cells within islets of Langerhans. Diabetes 37, 616–621 (1988)
Gu, C. et al. Pancreatic beta cells require NeuroD to achieve and maintain functional maturity. Cell Metab. 11, 298–310 (2010)
Blum, B. et al. Functional β-cell maturation is marked by an increased glucose threshold and by expression of urocortin 3. Nat. Biotechnol. 30, 261–264 (2012)
In’t Veld, P. & Marichal, M. Microscopic anatomy of the human islet of Langerhans. Adv. Exp. Med. Biol. 654, 1–19 (2010)
Meda, P., Perrelet, A. & Orci, L. Increase of gap junctions between pancreatic B-cells during stimulation of insulin secretion. J. Cell Biol. 82, 441–448 (1979)
Wang, Y. & Nathans, J. Tissue/planar cell polarity in vertebrates: new insights and new questions. Development 134, 647–658 (2007)
Wallingford, J. B. Planar cell polarity and the developmental control of cell behavior in vertebrate embryos. Annu. Rev. Cell Dev. Biol. 28, 627–653 (2012)
Eiraku, M. et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 472, 51–56 (2011)
McCracken, K. W. et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 516, 400–404 (2014)
Orci, L. et al. A morphological basis for intercellular communication between α- and β-cells in the endocrine pancreas. J. Clin. Invest. 56, 1066–1070 (1975)
Lange, A. et al. Fltp(T2AiCre): a new knock-in mouse line for conditional gene targeting in distinct mono- and multiciliated tissues. Differentiation 83, S105–S113 (2012)
Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L. & Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007)
Rieck, S. & Kaestner, K. H. Expansion of β-cell mass in response to pregnancy. Trends Endocrinol. Metab. 21, 151–158 (2010)
Speier, S. et al. Noninvasive in vivo imaging of pancreatic islet cell biology. Nat. Med. 14, 574–578 (2008)
Golson, M. L., Misfeldt, A. A., Kopsombut, U. G., Petersen, C. P. & Gannon, M. High fat diet regulation of β-cell proliferation and β-cell mass. Open Endocrinol. J. 4, (2010). 10.2174/1874216501004010066
Kitsou-Mylona, I., Burns, C. J., Squires, P. E., Persaud, S. J. & Jones, P. M. A role for the extracellular calcium-sensing receptor in cell-cell communication in pancreatic islets of Langerhans. Cell. Physiol. Biochem. 22, 557–566 (2008)
Schulze, T., Morsi, M., Bruning, D., Schumacher, K. & Rustenbeck, I. Different responses of mouse islets and MIN6 pseudo-islets to metabolic stimulation: a note of caution. Endocrine 51, 440–447 (2016)
Granot, Z. et al. LKB1 regulates pancreatic β cell size, polarity, and function. Cell Metab. 10, 296–308 (2009)
Kalwat, M. A. & Thurmond, D. C. Signaling mechanisms of glucose-induced F-actin remodeling in pancreatic islet β cells. Exp. Mol. Med. 45, e37 (2013)
Staiger, H. et al. Common genetic variation in the human FNDC5 locus, encoding the novel muscle-derived ‘browning’ factor irisin, determines insulin sensitivity. PLoS One 8, e61903 (2013)
Gao, T. et al. Pdx1 maintains β cell identity and function by repressing an α cell program. Cell Metab. 19, 259–271 (2014)
Wang, Z., York, N. W., Nichols, C. G. & Remedi, M. S. Pancreatic β cell dedifferentiation in diabetes and redifferentiation following insulin therapy. Cell Metab. 19, 872–882 (2014)
Talchai, C., Xuan, S., Lin, H. V., Sussel, L. & Accili, D. Pancreatic β cell dedifferentiation as a mechanism of diabetic β cell failure. Cell 150, 1223–1234 (2012)
Jonas, J. C. et al. Chronic hyperglycemia triggers loss of pancreatic β cell differentiation in an animal model of diabetes. J. Biol. Chem. 274, 14112–14121 (1999)
Szot, G. L., Koudria, P. & Bluestone, J. A. Murine pancreatic islet isolation. J. Vis. Exp. 2007, 255 (2007)
Ravassard, P. et al. A genetically engineered human pancreatic β cell line exhibiting glucose-inducible insulin secretion. J. Clin. Invest. 121, 3589–3597 (2011)
Miyazaki, J. et al. Establishment of a pancreatic β cell line that retains glucose-inducible insulin secretion: special reference to expression of glucose transporter isoforms. Endocrinology 127, 126–132 (1990)
Chmelova, H. et al. Distinct roles of β-cell mass and function during type 1 diabetes onset and remission. Diabetes 64, 2148–2160 (2015)
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012)
Dalton, A. J. A chrome-osmium fixative for electron microscopy. Anat. Rec. 121, 281 (1955)
Feuchtinger, A. et al. Image analysis of immunohistochemistry is superior to visual scoring as shown for patient outcome of esophageal adenocarcinoma. Histochem. Cell Biol. 143, 1–9 (2015)
Gegg, M. et al. Flattop regulates basal body docking and positioning in mono- and multiciliated cells. Elife 3, e03842 (2014)
Wu, Y. et al. Growth hormone receptor regulates β cell hyperplasia and glucose-stimulated insulin secretion in obese mice. J. Clin. Invest. 121, 2422–2426 (2011)
Stefan, N. et al. Polymorphisms in the gene encoding adiponectin receptor 1 are associated with insulin resistance and high liver fat. Diabetologia 48, 2282–2291 (2005)
Matsuda, M. & DeFronzo, R. A. Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care 22, 1462–1470 (1999)
Fadista, J. et al. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proc. Natl Acad. Sci. USA 111, 13924–13929 (2014)
Robinson, M. D. & Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 (2010)
R Computing Team. R: A Language and Environment for Statistical Computing. (R Foundation for Statistical Computing, Vienna, Austria, 2014)
Rainer, J., Sanchez-Cabo, F., Stocker, G., Sturn, A. & Trajanoski, Z. CARMAweb: comprehensive R- and bioconductor-based web service for microarray data analysis. Nucleic Acids Res. 34, W498–W503 (2006)
Acknowledgements
We thank A. Raducanu, A. Böttcher and E. Schlüssel for comments and discussions; A. Theis, B. Vogel and K. Diemer for technical support; the Human Tissue Laboratory (HTL) of Lund University Diabetes Centre (LUDC) for high-quality RNA sequencing expression data from human pancreatic islets donors; to InSphero for human microislets; the European Consortium for Islet Transplantation (ECIT) for human islets; and R. Scharfmann for the EndoC-β H1 cell line. The human islet research was supported by a Strategic Research Grant from the Swedish Research Council (2009-1039). A.M. was funded by the Helmholtz post-doctoral fellowship program. This work was supported by an Emmy-Noether Fellowship, the European Union (ERC starting grant Ciliary Disease) and the HumEn project from the European Union's Seventh Framework Programme for Research, Technological Development and Demonstration under grant agreement No. 602587 (http://www.hum-en.eu/). This work was funded (in part) for H.L. and J.B. by the Helmholtz Alliance ICEMED – Imaging and Curing Environmental Metabolic Diseases, through the Initiative and Networking Fund of the Helmholtz Association. We thank the Helmholtz Society, Helmholtz Portfolio Theme 'Metabolic Dysfunction and Common Disease, German Research Foundation and German Center for Diabetes Research (DZD e.V.) for financial support. This work was supported with funds for S.S. from the Emmy-Noether Program, the Center for Regenerative Therapies Dresden-DFG Research Center for Regenerative Therapies Dresden, Cluster of Excellence (CRTD), the DFG-Collaborative Research Center/Transregio 127 and the German Ministry for Education and Research to the German Centre for Diabetes Research and to the Network of Competence for Diabetes.
Author information
Authors and Affiliations
Contributions
E.B., A.M. and M.G. conceived and performed the experiments and wrote the manuscript. M.B. established the 3D in vitro cultures. S.S.R. and E.B. helped to perform human islet experiments. N.M., C.L., H.Z., M.J., M.T. and J.G. conceived and performed mitochondria analysis. M.I. and J.B. performed the microarray analysis. M.A. and A.F. analysed mitochondria by TEM. F.M., H.S. and H-U.H. performed the SNP analysis of FLTP in humans. O.K. provided human islets and N.O. provided RNA sequencing data. R.W-S. supervised and trained E.B. in human epidemiology and genetics. H.C., J.A.C. and S.S. performed the islet transplantation and analyzed the data set. H.L. conceived the study and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
H.L., M.G., E.B. and A.M. are inventors of a European Patent (PCT/EP2015/056664) based on this work.
Additional information
Reviewer Information Nature thanks C. Wollheim and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 1 FVR expression is induced in compacted islet structures and correlates with high Nkx6.1 expression.
a, Overview of a cleared whole-mount pancreas from an FltpZV/+ mouse at E18.5, stained with anti-Nkx6.1 and anti-GFP antibodies and scanned with a laser confocal microscope. b–e, In compacted islets (dotted line), high FVR expression correlates with high Nkx6.1 expression. f–i, In cord-like structures of endocrine cells that left the ductal epithelium (dotted line), low Nkx6.1 expression correlates with low FVR expression. j, Quantification of the mean fluorescent intensity of Venus (FVR)+ and Nkx6.1+ cells in compacted and cord-like structures (n (mice) = 3, 5 cord-like structures, 6 compacted islets; ***P = 0.0005, two sided unpaired t-test). k, Quantification of Nkx6.1 (red) and Venus (green) single-positive cells as well as all Nkx6.1–Venus double-positive cells (yellow) in compacted and cord-like structures (n (mice) = 3, cord-like structures 570 cells, compacted islets 1,827 cells; **P = 0.0064 for Venus, **P = 0.0041 for Nkx6.1, *P = 0.0182 for Venus–Nkx6.1, two-sided unpaired t-test). Scale bars, 100 μm (a), 20 μm (b–i). Data represent mean ± s.e.m.
Extended Data Figure 2 FVR− and FVR+ β-cell subpopulations exhibit differential proliferative capacity and inter-islet heterogeneity in the head and tail of the pancreas.
a, b, LSM images showing FVR, Nkx6.1 and EdU (a) and quantification (b) of EdU+ in FVR− and FVR+ β-cell subpopulations in control and pregnant (G15.5) mice (green/red arrow heads indicate proliferating FVR+/FVR− β-cells. n (control) = 4 (8,500 Nkx6.1+ cells), n (pregnant) = 4 (10,938 Nkx6.1+ cells); **P (Ctrl) = 0.0049, **P (pregnant) = 0.0059, two-sided unpaired t-test). c, Scatter plot of FVR+ β-cell distribution relative to number of β-cells per islet in FltpZV/+ mice (n (mice) = 10, n (islets) = 512). d, Quantification of islet size distribution in head and tail of FltpZV/+ pancreas sections (n (mice) = 4, n (islets) = 299; *P (20–50) = 0.0419, *P (>100) = 0.0238, two-sided unpaired t-test). e, f, LSM images (e) and quantification (f) of FVR+ β-cell distribution in the pancreas head and tail (n (mice) = 4, n (islets) = 299, n (Nkx6.1+ cells) = 19,054; ****P < 0.0001, two-sided unpaired t-test). g, Quantification of EdU incorporation among FVR− and FVR+ β-cell subpopulations relative to number of β-cells per islet in pregnant (G15.5) FltpZV/+ mice (n (mice) = 4, n (islets) = 196, n (Nkx6.1+ cells) = 10,938). h, LSM images showing Nkx6.1-, p27- and FVR-expressing cells in islets of FltpZV/+ pregnant (G15.5) mice. i, Quantification of p27 and Venus fluorescent intensity shows that p27-high cells have a higher Venus fluorescent intensity than p27-low cells in pregnant (G15.5) mice (n (mice) = 3, n (p27 and Venus positive cells) = 4,204). j, Quantification of Venus and EdU fluorescent intensity shows that EdU-positive cells have lower Venus fluorescent intensity than EdU-negative cells in pregnant (G15.5) mice (n (mice) = 3, n (p27+, EdU+ and Nkx6.1+ cells) = 75, n (p27+ EdU− Nkx6.1+ cells) = 4,845). Scale bars, 50 μm (a, e, h). Data represent mean ± s.e.m.
Extended Data Figure 3 Validation of the sorting scheme and molecular and cellular analysis of FVR− and FVR+ endocrine subpopulations.
a, b, Quantification (a) and LSM images (b) of FACS-sorted FVR populations for the microarray analysis show equal numbers of endocrine cell lineages among the subpopulations (n (DAPI+ cells) = 789). c, Detection and purification of four distinct endocrine subpopulations (I–IV) on the basis of granularity and Venus expression. d, e, Quantification (d) and LSM images (e) of sorted FVR subpopulations I and II confirms high enrichment of Nkx6.1+ β-cells whereas III and IV exhibit high enrichment of glucagon+ α-cells (n (DAPI+ cells) = 3,275). f, Expression analysis of endocrine hormones and the exocrine marker Amy2a3 reveals minimal contamination with other endocrine cell types (n (independent experiments) = 6 (FVR+) and 5 (FVR−)). g, h, Validation of the microarray analysis by real-time qPCR of β-cell-enriched subpopulations (I, II) for β-cell maturation, mitochondria and metabolic signalling pathway genes (Atp5b, Hadh, Wnt4, Grb10, Wnt5b, Ins1, GFP, Fltp, Pcsk1, Gipr, Mafa: n (independent experiments) = 6; Bace2, Alpk1, Npy: n (independent experiments) = 5, Slc2a2: n (independent experiments) = 3; *P < 0.05, **P < 0.01, ***P < 0.001, two-sided unpaired t-test). i, Real-time qPCR of mtDNA (S12) versus nDNA (HBB) from β-cell enriched populations (I, II) (n (independent experiments) = 5, expression in the FVR+ subpopulation normalized on each FVR− subpopulation, error bars represent c.i.). j, Scatter plot of regulated polarity genes among FVR endocrine cells in microarray analysis (thin lines mark 1.5-fold difference in expression, P < 0.05, limma t-test). Scale bars, 100 μm (b, e). Data represent mean ± s.e.m. except where stated.
Extended Data Figure 4 Transmission electron microscopy (TEM) analysis of Fltp− and Fltp+ β-cell subpopulations and quantification of mitochondria membrane length.
a, b, TEM images of immunogold-labelled β-cells of FltpT2AiCre/+;Gt(ROSA)26mTmG/+ mice reveal no structural differences in Golgi complex (a) or endoplasmic reticulum (b). c, d, TEM images and detected areas of the algorithm used for mitochondria analysis (c) and analysis of inner mitochondria membrane length normalized on the mitochondria area in FVR+ and FVR− β-cells (d) (n (independent experiments) = 2, n (FVR+ cells) = 14, n (FVR− cells) = 16, n (mitochondria) = 544). e, TEM images of mitochondria in FVR+ compared to mitochondria in FVR− β-cells. Scale bars, 500 nm (a (Golgi complex and gold staining), c, e), 200 nm (a (insulin granules), b). Data represent mean.
Extended Data Figure 5 Analysis of oxygen consumption rate (OCR), mitochondria network and GSIS in FVR+ and FVR− β-cells.
a, OCR of FACS-sorted β-cell-enriched FVR+ and FVR− subpopulations after Antimycin A subtraction. b, Quantification of absolute OCR rates of FACS-sorted β-cell-enriched FVR+ and FVR− subpopulations after Antimycin A subtraction. FVR+ n (technical replicates) = 9, FVR− n (technical replicates) = 4, for each technical replicate 3 × 104 cells were plated; endocrine cells were isolated from 9 mice; two-sided unpaired t-test. c, OCR of FACS-sorted β-cell-enriched FVR+ and FVR− subpopulations after Antimycin A subtraction and baseline normalization. d, Quantification of relative OCR of FACS-sorted β-cell-enriched FVR+ and FVR− subpopulations after Antimycin A subtraction and baseline normalization. FVR+ n (technical replicates) = 9, FVR− n (technical replicates) = 4, two-sided unpaired t-test. e, h, LSM pictures of immunohistochemistry of isolated FltpZV/+ mouse islets cultured under high (16.5 mM) and low (2 mM) glucose concentrations with single-cell reconstructions (Imaris) showing the reconstructed membrane (light brown), the nuclei (white, green and blue) and the mitochondria (red). f, g, i, j, Enlargement of a single Nkx6.1+ cell under high (f) and low glucose (i) and an Nkx6.1+ FVR+ cell under high (g) and low glucose (j) with reconstruction of the nucleus, the membrane and the mitochondria. k, Quantification of the mitochondrial morphology of Nkx6.1–FVR double-positive cells (yellow) and Nkx6.1 single-positive cells (white) under high and low glucose stimulation (n (Nkx6.1+ FVR+) = 9, n (Nkx6.1+ FVR− in high glucose) = 4, n (Nkx6.1+ FVR− in low glucose) = 6, from three islets for low glucose and two islets for high glucose). l, FACS-sorted and re-aggregated β-cell-enriched subpopulations reveal differences in GSIS and arginine depolarization (n (independent experiments) = 3, n (aggregates FVR+) = 16, n (aggregates FVR−) = 12; *P = 0.0206, **P = 0.007, two-sided unpaired t-test). Scale bars, 10 μm (e, h), 2 μm (f, j), 3 μm (g, i). Data represent mean ± s.e.m.
Extended Data Figure 6 Fltp genetic lineage tracing of endocrine subpopulation dynamics upon islet transplantation and HFD.
a, Maximum intensity projection of laser backscatter to determine the volume of the same transplanted FltpT2AiCre/+;Gt(ROSA)26mTmG/+ islet before (Pre-diet) and after 8 weeks of HFD. b, Quantification of the total islet volume (transplanted FltpT2AiCre/+;Gt(ROSA)26mTmG/+) in mice on normal diet and HFD (n (islets) = 25 (from 4 mice on normal diet), n (islets) = 34 (from 5 mice on HFD); ****P < 0.0001, Sidak’s multiple comparison test). c, Quantification of glucose levels during intraperitoneal GTT by AUC quantification (n (mice on normal diet) = 4, n (mice on HFD) = 5; *P = 0.0223, **P = 0.0064, Sidak’s multiple comparison test). d, Longitudinal quantification of the relative volumes of islet size and vessel volume during engraftment time (n (islets) = 15 (from 4 mice); ****P < 0.0001, ***P = 0.0027, two-sided unpaired t-test). e, Quantification of mG and mT endocrine cell numbers every 10 μm in a confocal z-stack during engraftment (n (islets) = 15 (from 4 mice); ****P < 0.0001, two-sided unpaired t-test). Scale bars, 50 μm (a). Data represent mean ± s.e.m.
Extended Data Figure 7 3D architecture and Wnt/PCP induce β-cell maturation in Min6 cells.
a, c–e, g, h, LSM images (a, e) and quantification (c, g (3 days culture), d, h, (7 days culture)) of Nkx6.1 and MafA mean fluorescent intensity in Min6 cells in 2D and 3D culture (c, n (independent experiments) = 3, ***P = 0.0002; d, n (independent experiments) = 4, *P = 0.0264; g, n (independent experiments) = 2; h, n (independent experiments) = 3; Sidak’s multiple comparison test). b, f, Fluorescent intensity of Nkx6.1 (b) and MafA (f) in a single Min6 cell in 2D and 3D culture. White lines in a and e indicate measured regions. i, j, Gene expression analysis of Min6 cells cultured in 2D and 3D culture by qPCR (n (independent experiments) = 4; *P < 0.05, **P < 0.01, ***P < 0.001, two-sided unpaired t-test). k, l, LSM images (k) and quantification (l) of Nkx6.1+ Min6 cells in floating 3D culture treated with or without Wnt5a (n (independent experiments) = 3; n (cells) = 5,419 (control), 2,419 (Wnt5a); **P = 0.0036, Sidak’s multiple comparison test). m, n, Western blot (m) and quantification (n) of Nkx6.1 and Fltp protein in Min6 after treatment for 6 days with or without Wnt5a in 2D or 3D culture (western blots were normalized on γ-tubulin expression and standardized on 2D control, n (independent experiments) = 5). Scale bars, 20 μm (a, e, k). Data represent mean ± s.e.m.
Extended Data Figure 8 Modulation of maturation and proliferation through WNT signalling in human microislets and EndoC-β H1 human β-cell line and SNP association with insulin secretion defects in the FLTP gene.
a–e, LSM images (a–d; proliferating cells marked by arrow heads) and quantification (e) of NKX6.1–EdU double-positive EndoC-β H1 cells treated with or without WNT3A, WNT4 or WNT5A (n (independent experiments) = 3, 1,617 cells counted; *P = 0.019, two-sided unpaired t-test). f, g, Western blot (f) and quantification (g) of pJNK in EndoC-β H1 treated for 4 h with or without WNT4 (n (independent experiments) = 4; *P = 0.027, two-sided paired t-test). h, Fold change in insulin secretion in response to high glucose in human microislets treated with or without WNT4 or WNT5A (3 human donors; Ctrl: n (microislets) = 23, WNT4: n (microislets) = 25, WNT5A: n (microislets) = 26; *P = 0.0423, two-sided unpaired t-test). i, RNA sequencing expression data for SLC2A2 in human pancreatic islets from healthy donors (NGT, n = 66), pre-diabetic donors (IGT, n = 21) and type 2 diabetic donors (T2D, n = 19; one-way ANOVA). j, k, The minor allele of FLTP rs75715534 was significantly associated with increased insulin secretion in lean subjects (P = 0.0035), but not in obese subjects (P = 0.07). Adjustment of the insulin secretion index AUC Cpep0–30/AUC Glc0–30 (n = 2,228) was achieved by multiple linear regression modelling with gender, age and OGTT-derived insulin sensitivity as confounding variables. Scale bars, 25 μm (a, d). Data represent mean ± s.e.m. (e, g, h); mean diamonds represent mean ± 95% c.i. (j, k).
Extended Data Figure 9 Knockout of Fltp has no effect on β-cell maturation or proliferation.
a–d, Single plane LSM images of cryosections from adult FltpZV/+ (a, c) and FltpZV/ZV (b, d) mice. Some Venus (green), Nkx6.1 (red) and MafA (white; a, b, white arrowheads) or Ucn3 (white; c, d, dashed lines) triple-positive cells are highlighted. e, Quantification of Venus, Nkx6.1 and Ucn3 (white) or MafA (black) triple-positive cells shows no difference in maturation status of β-cells in FltpZV/+ and FltpZV/ZV mice (FltpZV/+: n (mice) = 3, MafA 2,349 cells, Ucn3 1,212 cells; FltpZV/ZV: n (mice) = 3, MafA 867 cells, Ucn3 1,825 cells). f–i, LSM images of immunohistochemistry on cryosections from P11 and adult Fltp+/+ (f, P11; h, adult) and FltpZV/ZV (g, P11; i, adult) mice showing the proliferation of islet cells. j, k, Quantification of proliferating β- and non-β-cells in FltpZV/ZV and Fltp+/+ mice at P11 (j) (Fltp+/+: n (mice) = 4, 6,556 β-cells and 4,325 non-β-cells; FltpZV/ZV: n (mice) = 4, 6,709 β-cells and 5,868 non-β-cells) and in adults (k) (Fltp+/+: n (mice) = 7, 9,803 β-cells and 4,338 non-β-cells; FltpZV/ZV: n (mice) = 9, 14,957 β-cells and 7,955 non-β-cells). Scale bar, 20 μm (a), 10 μm (b), 5 μm (c, d), 15 μm (f–i). Data represent mean ± s.d.
Extended Data Figure 10 Analysis of the metabolic phenotype of FltpZV/ZV mice.
a, Quantification of β-cell number in islets of Langerhans from Fltp+/+ and FltpZV/ZV mice at P11 or adult (n (mice) = 3 (P11 Fltp+/+, 6,556 Nkx6.1+ cells and FltpZV/ZV, 6,791 Nkx6.1+ cells), n (mice) = 6 (adult Fltp+/+, 3,202 Nkx6.1+ cells) and n (mice) = 4 (adult FltpZV/ZV, 6,263 Nkx6.1+ cells)). b, Intraperitoneal GTT in FltpZV/ZV and Fltp+/+ mice (Fltp+/+ n (mice) = 7 and FltpZV/ZV n (mice) = 7; **P = 0.0022, two-sided unpaired t-test). c, d, Concentration of glucagon (c) and insulin (d) in pancreatic tissue from FltpZV/ZV and Fltp+/+ mice (Fltp+/+ n (mice) = 5 and FltpZV/ZV n (mice) = 5). e, ITT in FltpZV/ZV and Fltp+/+ mice (Fltp+/+ n (mice) = 5 and FltpZV/ZV n (mice) = 3). f, Plasma insulin level in fasted Fltp+/+ and FltpZV/ZV mice (Fltp+/+ n (mice) = 11 and FltpZV/ZV n (mice) = 10; *P = 0.0405, two-sided unpaired t-test). g, GSIS in isolated islets from Fltp+/+ and FltpZV/ZV mice (Fltp+/+ n (mice) = 5 and FltpZV/ZV n (mice) = 5; *P = 0.0263, one-way ANOVA, Bonferroni’s multiple comparison test). Data represent mean ± s.e.m.
Supplementary information
Supplementary Information
This file contains the Raw Data files for Figures 2k, 4h, 4j and Extended Data Figures 7m and 8f. (PDF 218 kb)
Supplementary Tables
This file contains Supplementary Tables 1-5. (PDF 1661 kb)
Source data
Rights and permissions
About this article
Cite this article
Bader, E., Migliorini, A., Gegg, M. et al. Identification of proliferative and mature β-cells in the islets of Langerhans. Nature 535, 430–434 (2016). https://doi.org/10.1038/nature18624
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature18624
This article is cited by
-
The role of wnt signaling in diabetes-induced osteoporosis
Diabetology & Metabolic Syndrome (2023)
-
Single-nucleus RNA sequencing of human pancreatic islets identifies novel gene sets and distinguishes β-cell subpopulations with dynamic transcriptome profiles
Genome Medicine (2023)
-
Integrated transcriptomics contrasts fatty acid metabolism with hypoxia response in β-cell subpopulations associated with glycemic control
BMC Genomics (2023)
-
Deficiency of transcription factor Nkx6.1 does not prevent insulin secretion in INS-1E cells
Scientific Reports (2023)
-
Multiparity increases the risk of diabetes by impairing the proliferative capacity of pancreatic β cells
Experimental & Molecular Medicine (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.