Abstract
Haematopoietic stresses mobilize haematopoietic stem cells (HSCs) from the bone marrow to the spleen and induce extramedullary haematopoiesis (EMH). However, the cellular nature of the EMH niche is unknown. Here we assessed the sources of the key niche factors, SCF (also known as KITL) and CXCL12, in the mouse spleen after EMH induction by myeloablation, blood loss, or pregnancy. In each case, Scf was expressed by endothelial cells and Tcf21+ stromal cells, primarily around sinusoids in the red pulp, while Cxcl12 was expressed by a subset of Tcf21+ stromal cells. EMH induction markedly expanded the Scf-expressing endothelial cells and stromal cells by inducing proliferation. Most splenic HSCs were adjacent to Tcf21+ stromal cells in red pulp. Conditional deletion of Scf from spleen endothelial cells, or of Scf or Cxcl12 from Tcf21+ stromal cells, severely reduced spleen EMH and reduced blood cell counts without affecting bone marrow haematopoiesis. Endothelial cells and Tcf21+ stromal cells thus create a perisinusoidal EMH niche in the spleen, which is necessary for the physiological response to diverse haematopoietic stresses.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Abdel-Wahab, O. I. & Levine, R. L. Primary myelofibrosis: update on definition, pathogenesis, and treatment. Annu. Rev. Med. 60, 233–245 (2009)
Cheshier, S. H., Prohaska, S. S. & Weissman, I. L. The effect of bleeding on hematopoietic stem cell cycling and self-renewal. Stem Cells Dev. 16, 707–718 (2007)
Bennett, M., Pinkerton, P. H., Cudkowicz, G. & Bannerman, R. M. Hemopoietic progenitor cells in marrow and spleen of mice with hereditary iron deficiency anemia. Blood 32, 908–921 (1968)
Nakada, D. et al. Oestrogen increases haematopoietic stem-cell self-renewal in females and during pregnancy. Nature 505, 555–558 (2014)
Fowler, J. H. & Nash, D. J. Erythropoiesis in the spleen and bone marrow of the pregnant mouse. Dev. Biol. 18, 331–353 (1968)
Baldridge, M. T., King, K. Y., Boles, N. C., Weksberg, D. C. & Goodell, M. A. Quiescent haematopoietic stem cells are activated by IFN-γ in response to chronic infection. Nature 465, 793–797 (2010)
Burberry, A. et al. Infection mobilizes hematopoietic stem cells through cooperative NOD-like receptor and Toll-like receptor signaling. Cell Host Microbe 15, 779–791 (2014)
Morrison, S. J., Wright, D. E. & Weissman, I. L. Cyclophosphamide/granulocyte colony-stimulating factor induces hematopoietic stem cells to proliferate prior to mobilization. Proc. Natl Acad. Sci. USA 94, 1908–1913 (1997)
Dutta, P. et al. Myocardial infarction accelerates atherosclerosis. Nature 487, 325–329 (2012)
Lowell, C. A., Niwa, M., Soriano, P. & Varmus, H. E. Deficiency of the Hck and Src tyrosine kinases results in extreme levels of extramedullary hematopoiesis. Blood 87, 1780–1792 (1996)
Freedman, M. H. & Saunders, E. F. Hematopoiesis in the human spleen. Am. J. Hematol. 11, 271–275 (1981)
Tavassoli, M. & Weiss, L. An electron microscopic study of spleen in myelofibrosis with myeloid metaplasia. Blood 42, 267–279 (1973)
Johns, J. L. & Christopher, M. M. Extramedullary hematopoiesis: a new look at the underlying stem cell niche, theories of development, and occurrence in animals. Vet. Pathol. 49, 508–523 (2012)
Kiel, M. J. et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121, 1109–1121 (2005)
Miwa, Y. et al. Up-regulated expression of CXCL12 in human spleens with extramedullary haematopoiesis. Pathology 45, 408–416 (2013)
Chow, A. et al. CD169+ macrophages provide a niche promoting erythropoiesis under homeostasis and stress. Nature Med. 19, 429–436 (2013)
Morita, Y. et al. Functional characterization of hematopoietic stem cells in the spleen. Exp. Hematol. 39, 351–359 (2011)
Ding, L. & Morrison, S. J. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature 495, 231–235 (2013)
Ding, L., Saunders, T. L., Enikolopov, G. & Morrison, S. J. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature 481, 457–462 (2012)
Zhou, B. O., Yue, R., Murphy, M. M., Peyer, J. G. & Morrison, S. J. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell 15, 154–168 (2014)
Acharya, A., Baek, S. T., Banfi, S., Eskiocak, B. & Tallquist, M. D. Efficient inducible Cre-mediated recombination in Tcf21 cell lineages in the heart and kidney. Genesis 49, 870–877 (2011)
Acar, M. et al. Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal. Nature 526, 126–130 (2015)
Akashi, K., Traver, D., Miyamoto, T. & Weissman, I. L. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 404, 193–197 (2000)
de Boer, J. et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 33, 314–325 (2003)
Sugiyama, T., Kohara, H., Noda, M. & Nagasawa, T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity 25, 977–988 (2006)
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature Neurosci. 13, 133–140 (2010)
DeFalco, J. et al. Virus-assisted mapping of neural inputs to a feeding center in the hypothalamus. Science 291, 2608–2613 (2001)
Becker, K., Jährling, N., Saghafi, S. & Dodt, H. U. Immunostaining, dehydration, and clearing of mouse embryos for ultramicroscopy. Cold Spring Harb. Protoc. 2013, 743–744 (2013)
Irizarry, R. A. et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 (2003)
Cesta, M. F. Normal structure, function, and histology of the spleen. Toxicol. Pathol. 34, 455–465 (2006)
Tronche, F. et al. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nature Genet. 23, 99–103 (1999)
Zhu, X., Bergles, D. E. & Nishiyama, A. NG2 cells generate both oligodendrocytes and gray matter astrocytes. Development 135, 145–157 (2008)
Zhu, X. et al. Age-dependent fate and lineage restriction of single NG2 cells. Development 138, 745–753 (2011)
Logan, M. et al. Expression of Cre recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis 33, 77–80 (2002)
Rivers, L. E. et al. PDGFRA/NG2 glia generate myelinating oligodendrocytes and piriform projection neurons in adult mice. Nature Neurosci. 11, 1392–1401 (2008)
Cuttler, A. S. et al. Characterization of Pdgfrb-Cre transgenic mice reveals reduction of ROSA26 reporter activity in remodeling arteries. Genesis 49, 673–680 (2011)
Holtwick, R. et al. Smooth muscle-selective deletion of guanylyl cyclase-A prevents the acute but not chronic effects of ANP on blood pressure. Proc. Natl Acad. Sci. USA 99, 7142–7147 (2002)
Xin, H. B., Deng, K. Y., Rishniw, M., Ji, G. & Kotlikoff, M. I. Smooth muscle expression of Cre recombinase and eGFP in transgenic mice. Physiol. Genomics 10, 211–215 (2002)
Wendling, O., Bornert, J. M., Chambon, P. & Metzger, D. Efficient temporally-controlled targeted mutagenesis in smooth muscle cells of the adult mouse. Genesis 47, 14–18 (2009)
Acknowledgements
S.J.M. is a Howard Hughes Medical Institute Investigator, the Mary McDermott Cook Chair in Pediatric Genetics, the director of the Hamon Laboratory for Stem Cells and Cancer, and a Cancer Prevention and Research Institute of Texas Scholar. B.O.Z. was supported by a fellowship from the Leukemia and Lymphoma Society. We thank N. Loof and the Moody Foundation Flow Cytometry Facility, K. Correll and M. Gross for mouse colony management, and E. Olson and J. Mendell for providing Cre lines. This work was supported by the National Institutes of Health National Heart, Lung, and Blood Institute (HL097760).
Author information
Authors and Affiliations
Contributions
C.N.I. identified the cre alleles used in this study and analysed Scf and Cxcl12 conditional knockout mice after Cy+G-CSF treatment. B.O.Z. characterized the stromal cells in the spleen and analysed Scf and Cxcl12 conditional knockout mice after blood loss and pregnancy. M.A. generated and characterized the α-catulinGFP mice. M.M.M. analysed HSC localization in the spleen. Z.Z. performed all statistical analyses. J.R. examined spleen histology. C.N.I., B.O.Z., M.A., M.M.M. and S.J.M. designed the experiments and interpreted the results. C.N.I., B.O.Z. and S.J.M. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
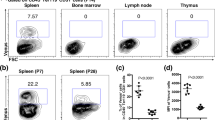
Extended Data Figure 1 Cy+21 d G-CSF treatment induces EMH in the spleen and deletion of Scf from LepR+ cells significantly reduces the number of HSCs in the bone marrow and the spleen after induction of EMH.
a,b, Staining with anti-laminin antibody distinguished the vasculature of red pulp (RP) from white pulp (WP). The red pulp and white pulp were marked by clusters of Ter119+ cells (red) and CD3+ cells (blue), respectively30. Dashed line depicts the boundary between red pulp and white pulp (representative images from 3 mice in 3 independent experiments). c, Spleen sections of the same magnification show the enlargement of the spleen after induction of EMH by Cy+21 d G-CSF. These are the same images as in Fig. 1a,d, adjusted to reflect the same magnification. d, e, Imaging of thick spleen sections from ScfGFP; Cxcl12DsRed mice after the induction of EMH by Cy+21 d G-CSF. e, High-magnification view of the boxed area in d. Dashed lines depict the boundaries between white pulp and red pulp. Arrow indicates the central arteriole in the white pulp around which stromal cells expressed Cxcl12-DsRed (representative images from 3 mice from 3 independent experiments). f, g, Haematoxylin and eosin (H&E) staining showing the increase in haematopoiesis in the spleen after induction of EMH using Cy+G-CSF (+EMH, g) as evidenced by the presence of megakaryocytes (arrows; n = 3 mice per condition from 3 independent experiments). h–n, Cy+G-CSF treatment significantly increased spleen cellularity (h), as well as the numbers of HSCs (i), MEPs (j), frequencies of colony-forming progenitors (k), numbers of Ter119+ erythroid cells (l) and Gr-1+Mac-1+ myeloid cells (m) in the spleen but not the number of B220+ or CD3+ lymphoid cells (n). The numbers of mice per treatment are shown in each bar of each panel. Each panel shows mean ± s.d. from five independent experiments. o, p, Scf-GFP (o) and Cxcl12-DsRed (p) fluorescence by spleen stromal cells before (−EMH) and after induction of EMH (+EMH) using Cy+G-CSF. q, r, The frequencies (q) and absolute numbers (r) of Scf-GFP+VE-cadherin+ endothelial cells and Scf-GFP+VE-cadherin− stromal cells significantly increased upon induction of EMH by Cy+21 d G-CSF (+EMH). s, Spleens from Leprcre; R26tdTomato; ScfGFP mice showed Tomato expression was primarily in the stromal cells of the white pulp. Although most Scf-GFP expression was in endothelial cells and perivascular stromal cells of the red pulp (Fig. 1a–d), some Scf-GFP+ stromal cells were in the white pulp, most of which appeared to express LepR. Dashed line depicts the boundary between red pulp and white pulp (representative images of 6 mice from 4 independent experiments). t, Flow cytometric analysis of enzymatically dissociated spleen cells from Leprcre; R26tdTomato; ScfGFP mice showed that only a small minority of non-endothelial Scf-GFP+ cells were positive for Tomato (n = 3 mice from 3 independent experiments). u, Tomato+CD45−Ter119− stromal cells in the spleens of Lepr-cre; R26tdTomato mice expressed PDGFR-α, PDGFR-β, Sca-1 and LepR (n = 3 mice from 3 independent experiments). v, Percentage of all CFU-F colonies formed by enzymatically dissociated spleen cells from Leprcre; R26tdTomato mice that expressed Tomato. Macrophage colonies were excluded by staining with anti-CD45 antibody (n = 4 mice from 3 independent experiments). w, Leprcre; Scffl/− mice had significantly fewer HSCs in the bone marrow than wild-type and Scffl/− controls before induction of EMH (n = 4 mice per genotype per time point mice from 4 independent experiments). NT, not treated. x, y, Leprcre; Scffl/− mice displayed significantly lower spleen cellularity (x) and HSC number (y) in the spleen than wild-type and Scffl/− controls after induction of EMH with cyclophosphamide plus 4 days of G-CSF. The numbers of mice per treatment are shown in each bar. Data represent mean ± s.d. from 4 independent experiments. h–n, q, r, The statistical significance of differences was assessed using two-tailed Student’s t-tests (***P < 0.001). w–y, The statistical significance of differences between genotypes was assessed using repeated measures one-way ANOVAs with Greenhouse–Geisser correction and Tukey’s multiple comparison tests with individual variances computed for each comparison. *P < 0.05, **P < 0.01, statistical significance relative to wild-type (Scf+/+). †P < 0.05, ††P < 0.01, statistical significance between Scf+/− and Leprcre; Scffl/−.
Extended Data Figure 2 Scf is expressed by most endothelial cells but not by Tcf21+ perivascular cells in the liver; Cy+21 d G-CSF does not significantly change the recombination pattern of Tcf21-Cre/ER in the spleen, bone marrow or liver.
a–i, To identify Cre alleles that recombine in spleen, but not bone marrow, stromal cells we assessed the gene expression profile of spleen Scf-GFP+VE-cadherin− stromal cells (Extended Data Table 1). Nestin, NG2 (also known as Cspg4) and Prx1 were low or undetectable (data not shown). Nestin-Cre31, NG2-Cre32, NG2-Cre/ER33, and Prx1-Cre34 did not recombine widely or specifically in Scf-GFP+ stromal cells in the spleen (data not shown). Pdgfra and Pdgfrb were expressed by spleen Scf-GFP+ stromal cells but neither Pdgfra-Cre/ER (ref. 35) nor Pdgfrb-Cre (ref. 36) recombined efficiently (data not shown). Sm22 (also known as Tagln), Myh11, Sma (also known as Acta2) and Tcf21 were significantly more highly expressed by spleen than bone marrow Scf-GFP+ stromal cells (Extended Data Table 1). Sm22-Cre (ref. 37), Myh11-Cre (ref. 38) and Sma-Cre/ER (ref. 39) recombined in few spleen Scf-GFP+ stromal cells (data not shown). However, Tcf21-Cre/ER recombined in perivascular stromal cells in the spleen but not bone marrow (Fig. 2). a–c, Under normal conditions, Scf-GFP was expressed by most VE-cadherin+ endothelial cells (arrowheads in a) but not by Tcf21+ stromal cells (arrows in a) in the liver (n =3 mice from 3 independent experiments). d, e, EMH induced by Cy+21 d G-CSF did not alter the general distribution (d) or perivascular localization (e) of Tomato+ cells in the spleens of Tcf21cre/ER; R26tdTomato mice as compared to normal mice (Fig. 2a, d). f, g, Tomato expression was undetectable in the bone marrow of Tcf21cre/ER; R26tdTomato mice after Cy+G-CSF treatment irrespective of whether the bone marrow was analysed by whole-mount imaging (f) or flow cytometry (g). h, i, EMH induced by Cy+G-CSF did not significantly change the frequency (h) or perivascular localization (i, arrows) of Tomato+ cells in the livers of Tcf21cre/ER; R26tdTomato mice. d–i, n =3 mice from 3 independent experiments.
Extended Data Figure 3 Deep imaging of HSCs in the spleen; deletion of Scf or Cxcl12 from Tcf21-expressing stromal cells in the spleen reduced peripheral blood cell counts but did not affect bone marrow haematopoiesis.
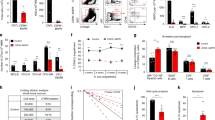
a, b, The vast majority of c-Kit+ haematopoietic progenitors localized adjacent to Tcf21-expressing stromal cells in the red pulp of the normal spleen (n = 3 mice from 3 independent experiments). c, d, Three-hundred-micrometre-thick sections of spleen before (c) and after optical clearing (d). e, f, Deep imaging of α-catulin-GFP+c-Kit+ HSCs in cleared spleen segments from Tcf21cre/ER; R26tdTomato; α-catulinGFP mice. A representative high-magnification image of an α-catulin-GFP+c-Kit+ HSC surrounded by Tomato+ stromal cells (e). f, Low-magnification view of a digitally reconstructed 300-μm-thick spleen fragment with α-catulin-GFP+c-Kit+ HSCs identified by large yellow spheres. Note that actual HSCs would be smaller than the yellow spheres but would not be visible at this magnification (n = 3 mice from 3 independent experiments). g–m, Tcf21cre/ER; Scffl/fl and Scffl/fl control mice were treated with tamoxifen then examined 1 month later without further treatment (not treated (NT)) or after treatment with cyclophosphamide plus 4, 8, or 21 days of G-CSF to induce EMH. Data show WBC (g), RBC (h) and PLT counts (i), numbers of CMPs (j), GMPs (k) and MEPs (l) in the bone marrow and numbers of GMPs in the spleen (m). n, Platelet counts of sham-operated and splenectomized mice that were treated with Cy+21 d G-CSF 1 month after surgery. o–u, Tcf21cre/ER; Cxcl12fll− mice and littermate controls (Cxcl12fl/− or Cxcl12+l−) were treated with tamoxifen then examined 1 month later without further treatment (NT) or after treatment with cyclophosphamide plus 4, 8, or 21 days of G-CSF to induce EMH. Data show WBC (o), RBC (p) and PLT counts (q), numbers of CMPs (r), GMPs (s) and MEPs (t) in the bone marrow and numbers of GMPs in the spleen (u). The numbers of mice per treatment are shown in each panel. All data reflect mean ± s.d. from 3 independent experiments. Two-tailed Student’s t-tests were used to assess statistical significance (*P < 0.05, ***P < 0.001).
Extended Data Figure 4 Conditional deletion of Scf or Cxcl12 with Tcf21-Cre/ER, or Scf with Vav1-Cre, does not significantly affect the frequency or morphology of stromal cells in the spleen, irrespective of EMH induction.
a–g, Irrespective of whether the mice were treated with Cy+21 d G-CSF, conditional deletion of Scf from Tcf21+ cells did not significantly change the frequency of VE-cadherin+ endothelial cells (a) or PDGFR-β+ perivascular stromal cells (b), Scf transcript levels in endothelial cells (c), or the morphology or density of blood vessels in the spleen (d–g). h–n, Irrespective of whether the mice were treated with Cy+G-CSF, conditional deletion of Cxcl12 from Tcf21+ cells did not significantly change the frequency of VE-cadherin+ endothelial cells (h) or PDGFR-β+ perivascular stromal cells (i), Scf transcript levels in endothelial cells or perivascular stromal cells (j), or the morphology or density of blood vessels in the spleen (k–n). o–u, Irrespective of whether the mice were treated with Cy+G-CSF, conditional deletion of Scf using Vav1-Cre did not significantly change the frequency of VE-cadherin+ endothelial cells (o) or PDGFR-β+ perivascular stromal cells (p), Scf transcript levels in perivascular stromal cells (q), or the morphology or density of blood vessels in the spleen (r–u). Scf transcript levels in flow cytometrically isolated cells were normalized to β-actin and then compared to whole spleen cells (c, j and q). The data reflect mean ± s.d. from 3 mice per genotype per condition in 3 independent experiments. Two-tailed Student’s t-tests were used to assess statistical significance.
Extended Data Figure 5 Vav1-Cre recombines efficiently and specifically in spleen endothelial cells but poorly in bone marrow or liver endothelial cells.
a, TomatohighCD45−Ter119− cells in Vav1-cre; tdTomato mice were uniformly positive for VE-cadherin and negative for PDGFR-β (n = 3 mice from 3 independent experiments). b, Vav1-Cre recombined in most spleen endothelial cells but in few bone marrow endothelial cells, irrespective of Cy+G-CSF treatment (+EMH). c, Scf transcript levels were significantly reduced in endothelial cells from the spleen but not from bone marrow or liver in Vav1-cre; Scffl/− mice as compared to Scffl/− mice. The Scf transcript level was normalized to β-actin. d, Western blot showed lower SCF protein levels in splenic endothelial cells from Vav1-cre; Scffl/− mice as compared to Scffl/− mice. SCF abundance was assessed relative to actin by Image J software (n = 3 mice per genotype from 3 independent experiments). e–h, In the bone marrow Vav1-Cre recombined in a minority of endothelial cells, including some sinusoidal (arrows in h) and some arteriolar (arrowheads in h) endothelial cells, that expressed little Scf-GFP by flow cytometry (f, g). The data reflect mean ± s.d. from 3 mice per genotype in 3 independent experiments. i–k, Vav1-Cre recombined inefficiently in liver endothelial cells. Most Tomato+ cells in the liver of Vav1-cre; R26tdTomato; ScfGFP mice were VE-cadherin+ and Scf-GFP+ (i; arrows in k) but these cells accounted for only 26 ± 4.2% of Scf-GFP+ cells by flow cytometry (i, j) and confocal microscopy (k, n = 3 mice from 3 independent experiments). Two-tailed Student’s t-tests were used to assess statistical significance.
Extended Data Figure 6 EMH induced by Cy+G-CSF does not significantly change the recombination pattern of Vav1-Cre in the spleen, bone marrow, or liver but deletion of Scf from endothelial cells in spleens with EMH reduces blood cell counts without affecting bone marrow haematopoiesis.
a, b, After EMH induced by Cy+21 d G-CSF, Vav1-Cre-recombined cells were predominantly in the red pulp (a) and co-localized with VE-cadherin+ cells (b) in the spleen. c–f, After EMH induced by Cy+21 d G-CSF, Vav1-Cre-recombined cells remained rare in the bone marrow (c, d) and liver (e, f; n = 3 mice from 3 independent experiments). g–s, Vav1-cre; Cxcl12fl/− mice and Cxcl12fll− controls were treated with Cy+4–21 d G-CSF to induce EMH. Data show WBC (g), RBC (h), and platelet (i) counts, spleen cellularity (j) and numbers of HSCs (k), CMPs (l), GMPs (m) and MEPs (n) in the spleen as well as bone marrow cellularity (o), and numbers of HSCs (p), CMPs (q), GMPs (r) and MEPs (s) in one femur and one tibia. The data represent mean mean ± s.d. from 3 (Cy+4 d G-CSF treatment) and 5 (Cy+21 d G-CSF treatment) independent experiments. The number of mice per treatment is indicated on each bar. Two-tailed Student’s t-tests were used to assess statistical significance. t–z, Vav1-cre; Scffl/− mice and Scffl/+, Scffl/− controls were treated with Cy+4–21 d G-CSF to induce EMH. Data show WBC (t), RBC (u), and platelet (PLT) (v) counts, numbers of CMPs (w), GMPs (x) and MEPs (y) in the bone marrow as well as numbers of GMPs in the spleen (z). Note that after 21 days of G-CSF both Scffl/− and Vav1-cre; Scffl/− mice showed significantly lower CMP numbers relative to Scffl/+ mice but their CMP numbers were not significantly different from each other (w), indicating that CMP numbers in the bone marrow were not influenced by Scf deletion from spleen endothelial cells. The data represent mean ± s.d. from 3 (no treatment (NT)), 3 (4 days), 3 (8 days), and 8 (21 days) independent experiments. The number of mice per treatment is indicated on each bar. The statistical significance of differences among genotypes was assessed using repeated measures one-way ANOVAs with Greenhouse–Geisser correction and Tukey’s multiple comparison tests with individual variances computed for each comparison. *P < 0.05, **P < 0.01, statistical significance relative to Scffl/+ controls. †P < 0.05, ††P < 0.01, statistical significance between Scffl/− and Vav1-cre; Scffl/−.
Extended Data Figure 7 Pregnancy induces EMH and the proliferation of endothelial cells and stromal cells in the spleen without significantly changing the recombination pattern of Tcf21-Cre/ER in the spleen, bone marrow or liver.
a–v, Pregnant female mice were at gestation day 18.5. a, b, H&E staining showed increased haematopoiesis in the spleens of pregnant mice (b) as evidenced by the presence of megakaryocytes (arrows; n = 3 mice per condition from 3 independent experiments). c–i, Pregnancy significantly increased spleen cellularity (c), as well as the numbers of HSCs (d), MEPs (e, f), Ter119+ erythroid cells (g) and Gr-1+Mac-1+ myeloid cells (h) in the spleen but not the number of B220+ or CD3+ lymphoid cells (i). j, k, During pregnancy, Scf-GFP was expressed by VE-cadherin+ endothelial cells and VE-cadherin− stromal cells (j) while Cxcl12-DsRed was expressed by a subset of the VE-cadherin−Scf-GFP+ stromal cells (j, k). l, Whole-mount imaging of a thick spleen section from a pregnant ScfGFP; Cxcl12DsRed mouse (representative images from 3 mice in 3 independent experiments). m, n, In the spleen, the numbers of Scf-GFP+ cells (m) and Cxcl12-DsRed+ cells (n) significantly increased upon bleeding. o, Endothelial and stromal cells in the spleen proliferated after bleeding. BrdU was administered to ScfGFP mice or Cxcl12DsRed mice for 18 days, beginning in pregnant mice after the plug was observed. The number of mice per treatment is indicated on each bar. Each panel shows mean ± s.d. from 3 independent experiments. Two-tailed Student’s t-tests were used to assess statistical significance (**P < 0.01, ***P < 0.001). p–r, Pregnancy did not alter the general distribution (p), perivascular localization (q) or surface marker expression (r; PDGFR-β+ and LepR−) of Tomato+ cells in the spleens of Tcf21cre/ER; R26tdTomato mice. s, t, Tomato expression remained undetectable in the bone marrow of pregnant Tcf21cre/ER; R26tdTomato mice. u, v, During pregnancy, Tcf21-Cre/ER recombined in rare perivascular cells in the liver. p–v, n = 3 mice per genotype from 3 independent experiments.
Extended Data Figure 8 Conditional deletion of Cxcl12 from Tcf21+ stromal cells impairs EMH in the spleens of pregnant mice without significantly affecting bone marrow haematopoiesis.
a–x, Four-to-six-month-old female mice that had been treated with tamoxifen at least 2 months before were mated with normal wild-type males. Normal females and pregnant females at gestation day 18.5 were analysed. a–d, Conditional deletion of Scf from Tcf21+ cells did not significantly affect the numbers of GMPs (a), CMPs (b), MEPs (c), Ter119+ (erythroid), Gr-1+Mac-1+ (myeloid), CD3+ (T) and B220+ (B) cells (d) in one femur or one tibia. e, f, Conditional deletion of Scf from Tcf21+ cells significantly reduced GMPs (e), Ter119+ erythrocytes and Gr-1+Mac-1+ myeloid cells (f) in the spleen. g, h, Conditional deletion of Scf from Tcf21+ cells did not significantly affect WBC (g) or platelet counts (h). i–n, Conditional deletion of Cxcl12 from Tcf21+ cells did not significantly affect bone marrow cellularity (i), or the numbers of HSCs (j), GMPs (k) CMPs (l), MEPs (m), Ter119+ (erythroid), Gr-1+Mac-1+ (myeloid), CD3+ (T) and B220+ (B) cells (n) in the bone marrow. o–w, Spleen cellularity (o) and numbers of HSCs (p), GMPs (q), CMPs (r), MEPs (s), Ter119+ (erythroid), Gr-1+Mac-1+ (myeloid), CD3+ (T) and B220+ (B) cells (t) in the spleen and WBC (u), platelet (v) and RBC counts (w) in the blood. x, Conditional deletion of Cxcl12 from Tcf21+ cells in the spleens of pregnant mothers did not significantly affect fetal mass. The numbers of mice per treatment are shown in each bar within each panel. Each panel shows mean ± s.d. from 3 independent experiments. a–w, The statistical significance of differences among genotypes was assessed using a repeated measures one-way ANOVA with Greenhouse–Geisser correction along with Tukey’s multiple comparison tests with individual variances x, The statistical significance of differences was assessed using a two-way ANOVA. *P < 0.05, **P < 0.01, ***P < 0.001, statistical significance relative to normal mice. †P < 0.05, ††P < 0.01, †††P < 0.001, statistical significance between Scf mutant mice and control mice after bleeding.
Extended Data Figure 9 Bleeding induces EMH and the proliferation of endothelial cells and stromal cells in the spleen without significantly changing the recombination pattern of Tcf21-Cre/ER in the spleen, bone marrow, or liver.
a, b, H&E staining showed an increase in haematopoiesis in the spleen after repeated bleeding (b; bled) as evidenced by the presence of megakaryocytes (arrows; n = 3 mice per condition from 3 independent experiments). c–i, Bleeding significantly increased spleen cellularity (c), as well as the numbers of HSCs (d), MEPs (e, f), and the numbers of Ter119+ erythroid cells (g) and Gr-1+Mac-1+ myeloid cells (h) in the spleen but not the number of B220+ or CD3+ lymphoid cells (i). j, k, After EMH induced by bleeding, Scf-GFP was expressed by VE-cadherin+ endothelial cells and VE-cadherin− stromal cells (j) while Cxcl12-DsRed was expressed by a subset of the VE-cadherin−Scf-GFP+ stromal cells (j, k). l, Whole-mount imaging of a thick spleen section from a ScfGFP; Cxcl12DsRed mouse after bleeding (representative images from 3 mice in 3 independent experiments). m, n, The numbers of Scf-GFP+ cells (m) and Cxcl12-DsRed+ cells (n) significantly increased upon bleeding. o, Endothelial and stromal cells in the spleen proliferated after bleeding. BrdU was administered to ScfGFP mice or Cxcl12DsRed mice for 15 days beginning after the first bleeding. The numbers of mice per treatment are shown in each bar in each panel. Each panel shows mean ± s.d. from three independent experiments. Two-tailed Student’s t-tests were used to assess statistical significance (**P < 0.01, ***P < 0.001). p–r, Bleeding did not alter the general distribution (p), perivascular localization (q) or surface marker expression (r; PDGFR-β+ and LepR−) of Tomato+ cells in the spleens of Tcf21cre/ER; R26tdTomato mice. s, t, Tomato expression remained undetectable in bone marrow from Tcf21cre/ER; R26tdTomato mice after bleeding. u, v, After bleeding, Tcf21-Cre/ER recombined only in rare perivascular cells in the liver (p–v; n = 3 mice per genotype from 3 independent experiments).
Extended Data Figure 10 Blood loss does not significantly change the recombination pattern of Vav1-Cre in the spleen, bone marrow, or liver; conditional deletion of Cxcl12 from Tcf21+ spleen stromal cells in bled mice impairs EMH in the spleen without significantly affecting bone marrow haematopoiesis.
a–e′,Four-to-six-month-old mice with the indicated genetic backgrounds were repeatedly bled over a 2-week period. a–h, After EMH induced by blood loss, Vav1-Cre recombined efficiently in VE-cadherin+ endothelial cells in the red pulp of the spleen (a–c) but poorly in the bone marrow (d–f) and liver (g, h), similar to what we observed under normal conditions (see Fig. 4a–c and Extended Data Fig. 5b) (a–h; n = 3 mice from 3 independent experiments). i–n, Conditional deletion of Scf using Tcf21-Cre/ER and/or Vav1-Cre did not significantly affect the numbers of GMPs (i), CMPs (j), MEPs (k), Ter119+ (erythroid), Gr-1+Mac-1+ (myeloid), CD3+ (T) and B220+ (B) cells (l) in the bone marrow or WBC (m) or platelet counts in the blood (n). o, p, Conditional deletion of Scf using Tcf21-Cre/ER and/or Vav1-Cre significantly reduced GMPs (o), Ter119+ erythrocytes and Gr-1+Mac-1+ myeloid cells (p) in the spleen. i–p, Data represent mean ± s.d. from 3 independent experiments. q–v, Conditional deletion of Cxcl12 from Tcf21+ spleen cells did not significantly affect bone marrow cellularity (q), or the numbers of HSCs (r), GMPs (s) CMPs (t), MEPs (u), Ter119+ (erythroid), Gr-1+Mac-1+ (myeloid), CD3+ (T) and B220+ (B) cells (v) in one femur and one tibia from bled mice. w–b′, Conditional deletion of Cxcl12 from Tcf21+ spleen cells significantly reduced spleen cellularity (w), and the numbers of MEPs (a′) and erythroid cells (b′) in the spleens of bled mice. Conditional deletion of Cxcl12 from Tcf21+ spleen cells did not significantly affect the numbers of HSCs (x), GMPs (y), or CMPs (z) in the spleens of bled mice. c′–e′, Conditional deletion of Cxcl12 from Tcf21+ spleen cells significantly reduced RBC (c′) but not WBC (d′) or platelet counts (e′) in the blood of mice that had been repeatedly bled. q–e′, Data represent mean ± s.d. from 3 independent experiments. The numbers of mice per treatment are shown in each bar in each panel. Statistical significance of differences among genotypes was assessed using a repeated measures one-way ANOVA with Greenhouse–Geisser correction along with Tukey’s multiple comparison tests with individual variances. *P < 0.05, **P < 0.01, ***P < 0.001, statistical significance relative to normal mice. †P < 0.05, ††P < 0.01, †††P < 0.001, statistical significance between Scf mutant mice and control mice after bleeding.
Supplementary information
HSCs are closely associated with Tcf21-Cre/ER-expressing stromal cells in the red pulp of the spleen
The video shows a 300 µm thick section of spleen from a Tcf21cre/ER ; R26tdTomato ; α-catulin-GFP mouse in which EMH was induced by treatment with Cy+21d G-CSF. The spleen section was stained with antibodies against c-kit (white), α-catulin-GFP (green), and DsRed (red), then cleared, imaged, and digitally reconstructed. HSCs are α-catulin-GFP+c-kit+. To show the spatial relationship between α-catulin-GFP+c-kit+ HSCs and Tomato+ cells all channels 20-30 µm beyond the spot of interest were occasionally masked. Note that the capsule on the margin of the spleen section is highly autofluorescent. (MP4 25025 kb)
Rights and permissions
About this article
Cite this article
Inra, C., Zhou, B., Acar, M. et al. A perisinusoidal niche for extramedullary haematopoiesis in the spleen. Nature 527, 466–471 (2015). https://doi.org/10.1038/nature15530
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature15530
This article is cited by
-
Extramedullary hematopoiesis in cancer
Experimental & Molecular Medicine (2024)
-
Skull bone marrow channels as immune gateways to the central nervous system
Nature Neuroscience (2023)
-
Splenic stromal niches in homeostasis and immunity
Nature Reviews Immunology (2023)
-
Hematopoiesis in the spleen after engraftment in unrelated cord blood transplantation evaluated by 18F-FLT PET imaging
International Journal of Hematology (2023)
-
Splenic red pulp macrophages provide a niche for CML stem cells and induce therapy resistance
Leukemia (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.