Abstract
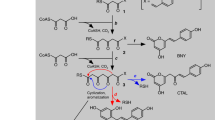
Specialized metabolic enzymes biosynthesize chemicals of ecological importance, often sharing a pedigree with primary metabolic enzymes1. However, the lineage of the enzyme chalcone isomerase (CHI) remained unknown. In vascular plants, CHI-catalysed conversion of chalcones to chiral (S)-flavanones is a committed step in the production of plant flavonoids, compounds that contribute to attraction, defence2 and development3. CHI operates near the diffusion limit with stereospecific control4,5. Although associated primarily with plants, the CHI fold occurs in several other eukaryotic lineages and in some bacteria. Here we report crystal structures, ligand-binding properties and in vivo functional characterization of a non-catalytic CHI-fold family from plants. Arabidopsis thaliana contains five actively transcribed genes encoding CHI-fold proteins, three of which additionally encode amino-terminal chloroplast-transit sequences. These three CHI-fold proteins localize to plastids, the site of de novo fatty-acid biosynthesis in plant cells. Furthermore, their expression profiles correlate with those of core fatty-acid biosynthetic enzymes, with maximal expression occurring in seeds and coinciding with increased fatty-acid storage in the developing embryo. In vitro, these proteins are fatty-acid-binding proteins (FAPs). FAP knockout A. thaliana plants show elevated α-linolenic acid levels and marked reproductive defects, including aberrant seed formation. Notably, the FAP discovery defines the adaptive evolution of a stereospecific and catalytically ‘perfected’ enzyme6 from a non-enzymatic ancestor over a defined period of plant evolution.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Hartmann, T. From waste products to ecochemicals: fifty years research of plant secondary metabolism. Phytochemistry 68, 2831–2846 (2007)
Ferrer, J.-L., Austin, M. B., Stewart, C., Jr & Noel, J. P. Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol. Biochem. 46, 356–370 (2008)
Peer, W. A. & Murphy, A. S. Flavonoids and auxin transport: modulators or regulators? Trends Plant Sci. 21, 556–563 (2007)
Bednar, R. A. & Hadcock, J. R. Purification and characterization of chalcone isomerase from soybeans. J. Biol. Chem. 263, 9582–9588 (1988)
Jez, J. M. & Noel, J. P. Reaction mechanism of chalcone isomerase: pH dependence, diffusion control, and product binding differences. J. Biol. Chem. 277, 1361–1369 (2002)
Albery, W. J. & Knowles, J. R. Evolution of enzyme function and the development of catalytic efficiency. Biochemistry 15, 5631–5640 (1976)
Jez, J. M., Bowman, M. E. & Noel, J. P. Role of hydrogen bonds in the reaction mechanism of chalcone isomerase. Biochemistry 41, 5168–5176 (2002)
Jez, J. M., Bowman, M. E., Dixon, R. A. & Noel, J. P. Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase. Nature Struct. Biol. 7, 786–791 (2000)
Gensheimer, M. & Mushegian, A. Chalcone isomerase family and fold: no longer unique to plants. Protein Sci. 13, 540–544 (2004)
Shirley, B. W., Hanley, S. & Goodman, H. M. Effects of ionizing radiation on a plant genome: analysis of two Arabidopsis transparent testa mutations. Plant Cell 4, 333–347 (1992)
Oursel, D. et al. Identification and relative quantification of fatty acids in Escherichia coli membranes by gas chromatography/mass spectrometry. Rapid Commun. Mass Spectrom. 21, 3229–3233 (2007)
Niesen, F. H., Berglund, H. & Vedadi, M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nature Protocols 9, 2212–2221 (2007)
Mentzen, W. I. & Wurtele, E. S. Regulon organization of Arabidopsis . BMC Plant Biol. 8, 99 (2008)
Mentzen, W. I., Peng, J., Ransom, N., Nikolau, B. J. & Wurtele, E. S. Articulation of three core metabolic processes in Arabidopsis: fatty acid biosynthesis, leucine catabolism and starch metabolism. BMC Plant Biol. 8, 76–90 (2008)
Ruuska, S. A., Girke, T., Benning, C. & Ohlrogge, J. B. Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14, 1191–1206 (2002)
Falcone, D. L., Ogas, J. P. & Somerville, C. R. Regulation of membrane fatty acid composition by temperature in mutants of Arabidopsis with alterations in membrane lipid composition. BMC Plant Biol. 4, 17–31 (2004)
Murakami, Y., Tsuyama, M., Kobayashi, Y., Kodama, H. & Iba, K. Trienoic fatty acids and plant tolerance of high temperature. Science 287, 476–479 (2000)
Austin, M. B. & Noel, J. P. The chalcone synthase superfamily of type III polyketide synthases. Nat. Prod. Rep. 20, 79–110 (2003)
Hilson, P. et al. Versatile gene-specific tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genet. Res. 14, 2176–2189 (2004)
Bonaventure, G., Salas, J. J., Pollard, M. R. & Ohlrogge, J. B. Disruption of the FATB gene in Arabidopsis demonstrates an essential role of saturated fatty acids in plant growth. Plant Cell 15, 1020–1033 (2003)
DeLano, W. L. The PyMOL Molecular Graphics System. (DeLano Scientific, 2002)
Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Birney, E., Clamp, M. & Durbin, R. GeneWise and Genomewise. Genome Res. 14, 988–995 (2004)
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004)
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007)
Huelsenbeck, J. P. & Ronquist, F. MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17, 754–755 (2001)
Guindon, S., Lethiec, F., Duroux, P. & Gascuel, O. PHYML Online – a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33, W557–W559 (2005)
Jez, J. M., Ferrer, J. L., Bowman, M. E., Dixon, R. A. & Noel, J. P. Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase. Biochemistry 39, 890–902 (2000)
Doublié, S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 276, 523–530 (1997)
Niesen, F. H., Berglund, H. & Vedadi, M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nature Protocols 9, 2212–2221 (2007)
Leslie, A. G. W. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography No. 26. (1992)
Evans, P. R. Scaling and assessment of data quality. Acta Crystallogr. D 62, 72–82 (2005)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 26, 795–800 (1993)
Terwilliger, T. C. & Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D 55, 849–861 (1999)
Terwilliger, T. C. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. D 59, 38–44 (2003)
Vagin, A. & Teplyakov, A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 30, 1022–1025 (1997)
Sali, A. & Blundell, T. L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993)
Perrakis, A., Morris, R. & Lamzin, V. S. Automated protein model building combined with iterative structure refinement. Nature Struct. Biol. 6, 458–463 (1999)
Brunger, A. T. & Warren, G. L. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
McRee, D. E. XtalView/Xfit: a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125, 156–165 (1999)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Krissinel, E. & Henrick, K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D 60, 2256–2268 (2004)
Invitrogen. Gateway technology: a universal technology to clone DNA sequences for functional analysis and expression in multiple systems. Catalog nos. 12535-019 and 12535-027. http://wolfson.huji.ac.il/expression/gatewayman.pdf (2003)
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743 (1998)
Lohar, D. P., Shuller, K., Buzas, D. M., Gresshoff, P. M. & Stiller, J. Transformation of Lotus japonicum using the herbicide resistance bar gene as a selectable marker. J. Exp. Bot. 52, 1697–1702 (2001)
Li, L., Ilarslan, H., James, M. G., Myers, A. M. & Wurtele, E. S. Genome wide co-expression among the starch debranching enzyme genes AtISA1, AtISA2, and AtISA3 in Arabidopsis thaliana . J. Exp. Bot. 58, 3323–3342 (2007)
Earley, K. W. et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629 (2006)
Jefferson, R. A. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 5, 387–405 (1987)
Doyle, J. J. & Doyle, J. L. Isolation of plant DNA from fresh tissue. Focus 12, 13–15 (1990)
Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Annu. Rev. Biochem. 72, 248–254 (1976)
Markham, J. E., Li, J., Cahoon, E. B. & Jaworski, J. G. Separation and identification of major plant sphingolipid classes from leaves. J. Biol. Chem. 281, 22684–22694 (2006)
Moon, S. & Nikolau, B. J. Plantmetabolomics Platform2: fatty acids. http://www.plantmetabolomics.org (2009)
Perera, M. A. D. N., Dietrich, C. R., Meeley, R., Schnable, P. S. & Nikolau, B. J. in Advanced Research on Plant Lipids (eds Murata, M. et al.) 225–228 (Kluwer Academic, 2003)
Fatland, B. L., Nikolau, B. J. & Wurtele, E. S. Reverse genetic characterization of cytosolic acetyl-CoA generation by ATP-citrate lyase in Arabidopsis . Plant Cell 17, 182–203 (2005)
Davies, N. A. The New Automated Mass Spectrometry Deconvolution and Identification System (AMDIS) (ISAS, 1998)
Acknowledgements
We thank A. Perera, B. Nikolau, H. Ilarsan and J. Peng for technical training (to M.N.N.), J. Peng for the fap1-1 homozygote mutant line, D. Nettleton and H. Wang for statistical analysis of initial seed fatty-acid data, and Eric Scheeff for assistance in Bayesian phylogenetic analysis. This research was supported in part by a Fulbright Fellowship (to M.N.N.). This material is based in part upon work supported by the National Science Foundation under award number MCB-0645794 (to J.P.N.), EEC-0813570 (to E.S.W.), MCB-0951170 (to E.S.W.), and by National Cancer Institute award number CA14195 (to G.M.) and the Plant Sciences Institute at Iowa State University (to E.S.W). J.P.N. is an investigator with the Howard Hughes Medical Institute. Portions of this research were conducted at the Advanced Light Source, a national user facility operated by Lawrence Berkeley National Laboratory, on behalf of the US Department of Energy, Office of Basic Energy Sciences. The Berkeley Center for Structural Biology is supported in part by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences. We thank the staff at the Advanced Light Source for assistance with X-ray data collection. Any opinions, findings and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
Author information
Authors and Affiliations
Contributions
M.N.N. experimentally characterized the FAP genes in planta. M.E.B., R.N.P. and E.L. expressed, purified and crystallized proteins. L.L. designed genetics experiments and constructs. G.V.L. and F.P. performed fatty-acid binding analyses and solved the X-ray crystal structures. R.N.P. performed thermal-shift assays of fatty-acid binding. G.M. and E.L. performed phylogenetic and sequence analyses. J.P.N. designed the biochemical experiments; E.S.W. designed the bioinformatics and functional genomics experiments. The manuscript was written by R.N.P., G.V.L., M.N.N., J.P.N., G.M. and E.S.W.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-23 and Supplementary Tables 1-9. (PDF 6495 kb)
Supplementary Data 1
This file contains CHI-fold Family Multisequence Alignment data. (TXT 107 kb)
Supplementary Data 2
This file contains CHI-fold Family Sequences and Accession Numbers. (XLS 83 kb)
Rights and permissions
About this article
Cite this article
Ngaki, M., Louie, G., Philippe, R. et al. Evolution of the chalcone-isomerase fold from fatty-acid binding to stereospecific catalysis. Nature 485, 530–533 (2012). https://doi.org/10.1038/nature11009
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11009
This article is cited by
-
Flavonoids are involved in salt tolerance through ROS scavenging in the halophyte Atriplex canescens
Plant Cell Reports (2024)
-
Multi-omics analysis reveals the molecular basis of flavonoid accumulation in fructus of Gardenia (Gardenia jasminoides Ellis)
BMC Genomics (2023)
-
Molecular cloning and in silico analysis of chalcone isomerase from Polygonum minus
Molecular Biology Reports (2023)
-
Identification and characterization of unique 5-hydroxyisoflavonoid biosynthetic key enzyme genes in Lupinus albus
Plant Cell Reports (2022)
-
Palmitoylated acyl protein thioesterase APT2 deforms membranes to extract substrate acyl chains
Nature Chemical Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.