Abstract
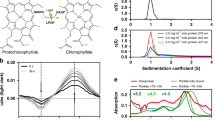
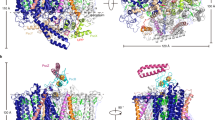
Photosynthetic organisms adopt two different strategies for the reduction of the C17 = C18 double bond of protochlorophyllide (Pchlide) to form chlorophyllide a, the direct precursor of chlorophyll a (refs 1–4). The first involves the activity of the light-dependent Pchlide oxidoreductase5,6,7,8,9, and the second involves the light-independent (dark-operative) Pchlide oxidoreductase10 (DPOR). DPOR is a nitrogenase-like enzyme consisting of two components, L-protein (a BchL dimer) and NB-protein (a BchN–BchB heterotetramer), which are structurally related to nitrogenase Fe protein and MoFe protein, respectively10,11. Here we report the crystal structure of the NB-protein of DPOR from Rhodobacter capsulatus at a resolution of 2.3 Å. As expected, the overall structure is similar to that of nitrogenase MoFe protein: each catalytic BchN–BchB unit contains one Pchlide and one iron–sulphur cluster (NB-cluster) coordinated uniquely by one aspartate and three cysteines. Unique aspartate ligation is not necessarily needed for the cluster assembly but is essential for the catalytic activity. Specific Pchlide-binding accompanies the partial unwinding of an α-helix that belongs to the next catalytic BchN–BchB unit. We propose a unique trans-specific reduction mechanism in which the distorted C17-propionate of Pchlide and an aspartate from BchB serve as proton donors for C18 and C17 of Pchlide, respectively. Intriguingly, the spatial arrangement of the NB-cluster and Pchlide is almost identical to that of the P-cluster and FeMo-cofactor in nitrogenase MoFe-protein, illustrating that a common architecture exists to reduce chemically stable multibonds of porphyrin and dinitrogen.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Protein Data Bank
Data deposits
Coordinates and structure factors for the structures reported here are available from the Protein Data Bank with accession codes of 3AEK (Pchlide-bound form), 3AEQ (anaerobic Pchlide-bound form), 3AER (Pchlide-free form), 3AES (selenomethionine-substituted Pchlide-free form), 3AET (D36C form) and 3AEU (D36A form).
Change history
06 May 2010
The position of the Mg in Fig. 3d was corrected.
References
Von Wettstein, D., Gough, S. & Kannangara, C. Chlorophyll biosynthesis. Plant Cell 7, 1039–1057 (1995)
Tanaka, R. & Tanaka, A. Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 58, 321–346 (2007)
Rüdiger, W. in Chlorophylls and Bacteriochlorophylls (eds Grimm, B., Porra, R. J., Rüdiger, W. & Scheer, H.) 189–200 (Biochem. Biophys. Funct. Appl. Ser. Adv. Photosynth. Respir. 25, Springer, 2006)
Masuda, T. & Fujita, Y. Regulation and evolution of chlorophyll metabolism. Photochem. Photobiol. Sci. 7, 1131–1149 (2008)
Wilks, H. M. & Timko, M. P. A light-dependent complementation system for analysis of NADPH:protochlorophyllide oxidoreductase: identification and mutagenesis of two conserved residues that are essential for enzyme activity. Proc. Natl Acad. Sci. USA 92, 724–728 (1995)
Lebedev, N. & Timko, M. Protochlorophyllide oxidoreductase B-catalyzed protochlorophyllide photoreduction in vitro: insight into the mechanism of chlorophyll formation in light-adapted plants. Proc. Natl Acad. Sci. USA 96, 9954–9959 (1999)
Masuda, T. & Takamiya, K. Novel insights into the enzymology, regulation and physiological functions of light-dependent protochlorophyllide oxidoreductase in angiosperms. Photosynth. Res. 81, 1–29 (2004)
Buhr, F. et al. Photoprotective role of NADPH:protochlorophyllide oxidoreductase A. Proc. Natl Acad. Sci. USA 105, 12629–12634 (2008)
Heyes, D. J. & Hunter, C. N. Making light work of enzyme catalysis: protochlorophyllide oxidoreductase. Trends Biochem. Sci. 30, 642–649 (2005)
Fujita, Y. & Bauer, C. E. in Chlorophylls and Bilins: Biosynthesis, Synthesis, and Degradation (eds Kadish, K., Smith, K. M. & Guilard, R.) 109–156 (Academic, 2003)
Fujita, Y. & Bauer, C. Reconstitution of light-independent protochlorophyllide reductase from purified BchL and BchN-BchB subunits. In vitro confirmation of nitrogenase-like features of a bacteriochlorophyll biosynthesis enzyme. J. Biol. Chem. 275, 23583–23588 (2000)
Reinbothe, S., Reinbothe, C., Apel, K. & Lebedev, N. Evolution of chlorophyll biosynthesis–the challenge to survive photooxidation. Cell 86, 703–705 (1996)
Kim, C.-H. & Rees, D. C. Structural models for the metal centers in the nitrogenase molybdenum-iron protein. Science 257, 1677–1682 (1992)
Einsle, O. et al. Nitrogenase MoFe-protein at 1.16 Å resolution: a central ligand in the FeMo-cofactor. Science 297, 1696–1700 (2002)
Schmid, B. et al. Structure of a cofactor-deficient nitrogenase MoFe protein. Science 296, 352–356 (2002)
Nomata, J., Ogawa, T., Kitashima, M., Inoue, K. & Fujita, Y. NB-protein (BchN-BchB) of dark-operative protochlorophyllide reductase is the catalytic component containing oxygen-tolerant Fe-S clusters. FEBS Lett. 582, 1346–1350 (2008)
Bröcker, M. J. et al. ATP-driven reduction by dark-operative protochlorophyllide oxidoreductase from Chlorobium tepidum mechanistically resembles nitrogenase catalysis. J. Biol. Chem. 283, 10559–10567 (2008)
Sarma, R. et al. Crystal structure of the L protein of Rhodobacter sphaeroides light-independent protochlorophyllide reductase with MgADP bound: a homologue of the nitrogenase Fe protein. Biochemistry 47, 13004–13015 (2008)
Beinert, H., Kennedy, M. & Stout, C. Aconitase as iron-sulfur protein, enzyme, and iron-regulatory protein. Chem. Rev. 96, 2335–2374 (1996)
Calzolai, L. et al. 1H NMR investigation of the electronic and molecular structure of the four-iron cluster ferredoxin from the hyperthermophile Pyrococcus furiosus. Identification of Asp 14 as a cluster ligand in each of the four redox states. Biochemistry 34, 11373–11384 (1995)
Imai, T. et al. Characterization and cloning of an extremely thermostable, Pyrococcus furiosus-type 4Fe ferredoxin from Thermococcus profundus. J. Biochem. 130, 649–655 (2001)
Page, C. C., Moser, C. C., Chen, X. & Dutton, P. L. Natural engineering principles of electron tunnelling in biological oxidation–reduction. Nature 402, 47–52 (1999)
Shah, V. K. & Brill, W. J. Isolation of an iron-molybdenum cofactor from nitrogenase. Proc. Natl Acad. Sci. USA 74, 3249–3253 (1977)
Nomata, J., Swem, L., Bauer, C. & Fujita, Y. Overexpression and characterization of dark-operative protochlorophyllide reductase from Rhodobacter capsulatus. Biochim. Biophys. Acta 1708, 229–237 (2005)
Nomata, J., Kitashima, M., Inoue, K. & Fujita, Y. Nitrogenase Fe protein-like Fe-S cluster is conserved in L-protein (BchL) of dark-operative protochlorophyllide reductase from Rhodobacter capsulatus. FEBS Lett. 580, 6151–6154 (2006)
Yamamoto, H., Nomata, J. & Fujita, Y. Functional expression of nitrogenase-like protochlorophyllide reductase from Rhodobacter capsulatus in Escherichia coli. Photochem. Photobiol. Sci. 7, 1238–1242 (2008)
Bricogne, G., Vonrhein, C., Flensburg, C., Schiltz, M. & Paciorek, W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. D 59, 2023–2030 (2003)
Abrahams, J. P. & Leslie, A. G. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D 52, 30–42 (1996)
Terwilliger, T. C. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 374, 22–37 (2003)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Collaborative. Computation Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Wallace, A. C., Laskowski, R. A. & Thornton, J. M. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 8, 127–134 (1995)
Mizoguchi, T. et al. Stereochemical determination of the unique acrylate moiety at the 17-position in chlorophylls-c from a diatom Chaetoseros calcitrans and its effect upon electronic absorption properties. Org. Biomol. Chem. 7, 2120–2126 (2009)
Acknowledgements
We thank K. Fukuyama, T. Omata and K. Terauchi for discussions; J. Peters and R. Cogdell for a critical reading of the manuscript; H. Yamamoto for establishing the functional overexpression system in E. coli; J. Harimoto for technical help in the sequence confirmation of a series of expression vectors; C. E. Bauer for donating the broad host-range vector pBBR1MSC2 and the triparental mating system for R. capsulatus; and the staff at the Photon Factory, KEK, Japan, for support during data collection. This work was supported by Grants-in-Aid for Scientific Research numbers 17687008 and 18054308 (G.K.), 19570036, 14COEA2 and 20200063 (Y.F.), 19750148 (T.M.) and 19350088 (H.T.) from the Japan Society for the Promotion of Science (JSPS), Precursory Research for Embryonic Science and Technology of the Japan Science and Technology Agency (JST) (Y.F.), the Kato Memorial Science Foundation (G.K.), the Toyoaki Scholarship Foundation and the Japan Securities Scholarship Foundation (Y.F.). N.M and J.N. grateful for fellowships from the JSPS for Japanese Junior Scientists (numbers 19010733 and 2110614).
Author information
Authors and Affiliations
Contributions
Purification of NB-protein and site-directed mutagenesis were conducted by J.N., K.E. and N.M.; the enzymatic assay of NB-protein was conducted by J.N.; the crystal structures of NB-protein were solved by N.M., T.S. and G.K.; chlorophyll c was prepared by T.M. and H.T.; G.K. and Y.F. contributed to the design of the experiments and writing the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-6 with legends, Supplementary Tables 1-2, a Supplementary Discussion and Supplementary References. (PDF 8590 kb)
Rights and permissions
About this article
Cite this article
Muraki, N., Nomata, J., Ebata, K. et al. X-ray crystal structure of the light-independent protochlorophyllide reductase. Nature 465, 110–114 (2010). https://doi.org/10.1038/nature08950
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature08950
This article is cited by
-
Chlorophyll biogenesis sees the light
Nature Plants (2021)
-
Consensus model of a cyanobacterial light-dependent protochlorophyllide oxidoreductase in its pigment-free apo-form and photoactive ternary complex
Communications Biology (2019)
-
Evolution of light-independent protochlorophyllide oxidoreductase
Protoplasma (2019)
-
Dark chlorophyll synthesis may provide a potential for shade tolerance as shown by a comparative study with seedlings of European larch (Larix decidua) and Norway spruce (Picea abies)
Trees (2018)
-
Structural and functional characterization of the hydrogenase-maturation HydF protein
Nature Chemical Biology (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.