Abstract
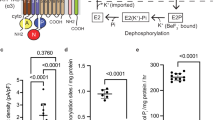
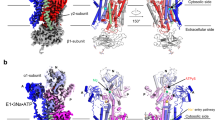
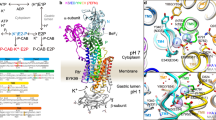
Sodium–potassium ATPase is an ATP-powered ion pump that establishes concentration gradients for Na+ and K+ ions across the plasma membrane in all animal cells by pumping Na+ from the cytoplasm and K+ from the extracellular medium1,2. Such gradients are used in many essential processes, notably for generating action potentials. Na+, K+-ATPase is a member of the P-type ATPases, which include sarcoplasmic reticulum Ca2+-ATPase and gastric H+, K+-ATPase, among others, and is the target of cardiac glycosides. Here we describe a crystal structure of this important ion pump, from shark rectal glands, consisting of α- and β-subunits and a regulatory FXYD protein3,4, all of which are highly homologous to human ones. The ATPase was fixed in a state analogous to E2·2K+·Pi, in which the ATPase has a high affinity for K+ and still binds Pi, as in the first crystal structure of pig kidney enzyme at 3.5 Å resolution5. Clearly visualized now at 2.4 Å resolution are coordination of K+ and associated water molecules in the transmembrane binding sites and a phosphate analogue (MgF42-) in the phosphorylation site. The crystal structure shows that the β-subunit has a critical role in K+ binding (although its involvement has previously been suggested6,7,8) and explains, at least partially, why the homologous Ca2+-ATPase counter-transports H+ rather than K+, despite the coordinating residues being almost identical.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Albers, R. W. Biochemical aspects of active transport. Annu. Rev. Biochem. 36, 727–756 (1967)
Post, R. L., Hegyvary, C. & Kume, S. Activation by adenosine triphosphate in the phosphorylation kinetics of sodium and potassium ion transport adenosine triphosphatase. J. Biol. Chem. 247, 6530–6540 (1972)
Mahmmoud, Y. A., Vorum, H. & Cornelius, F. Identification of a phospholemman-like protein from shark rectal glands. Evidence for indirect regulation of Na,K-ATPase by protein kinase c via a novel member of the FXYDY family. J. Biol. Chem. 275, 35969–35977 (2000)
Garty, H. & Karlish, S. J. Role of FXYD proteins in ion transport. Annu. Rev. Physiol. 68, 431–459 (2006)
Morth, J. P. et al. Crystal structure of the sodium–potassium pump. Nature 450, 1043–1049 (2007)
Lutsenko, S. & Kaplan, J. H. An essential role for the extracellular domain of the Na,K-ATPase β-subunit in cation occlusion. Biochemistry 32, 6737–6743 (1993)
Hasler, U., Crambert, G., Horisberger, J. D. & Geering, K. Structural and functional features of the transmembrane domain of the Na,K-ATPase β subunit revealed by tryptophan scanning. J. Biol. Chem. 276, 16356–16364 (2001)
Geering, K. The functional role of β subunits in oligomeric P-type ATPases. J. Bioenerg. Biomembr. 33, 425–438 (2001)
Schack, V. R. et al. Identification and function of a cytoplasmic K+ site of the Na+, K+ -ATPase. J. Biol. Chem. 283, 27982–27990 (2008)
Toyoshima, C., Nomura, H. & Tsuda, T. Lumenal gating mechanism revealed in calcium pump crystal structures with phosphate analogues. Nature 432, 361–368 (2004)
Toyoshima, C., Nakasako, M., Nomura, H. & Ogawa, H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Å resolution. Nature 405, 647–655 (2000)
Jacobsen, M. D., Pedersen, P. A. & Jorgensen, P. L. Importance of Na,K-ATPase residue alpha 1-Arg544 in the segment Arg544-Asp567 for high-affinity binding of ATP, ADP, or MgATP. Biochemistry 41, 1451–1456 (2002)
Toyoshima, C. & Mizutani, T. Crystal structure of the calcium pump with a bound ATP analogue. Nature 430, 529–535 (2004)
Sørensen, T. L., Møller, J. V. & Nissen, P. Phosphoryl transfer and calcium ion occlusion in the calcium pump. Science 304, 1672–1675 (2004)
Clausen, J. D., McIntosh, D. B., Vilsen, B., Woolley, D. G. & Andersen, J. P. Importance of conserved N-domain residues Thr441, Glu442, Lys515, Arg560, and Leu562 of sarcoplasmic reticulum Ca2+-ATPase for MgATP binding and subsequent catalytic steps. Plasticity of the nucleotide-binding site. J. Biol. Chem. 278, 20245–20258 (2003)
Murphy, A. J. & Hoover, J. C. Inhibition of the Na,K-ATPase by fluoride. Parallels with its inhibition of the sarcoplasmic reticulum CaATPase. J. Biol. Chem. 267, 16995–17000 (1992)
Jensen, A. M., Sørensen, T. L., Olesen, C., Møller, J. V. & Nissen, P. Modulatory and catalytic modes of ATP binding by the calcium pump. EMBO J. 25, 2305–2314 (2006)
Toyoshima, C. & Nomura, H. Structural changes in the calcium pump accompanying the dissociation of calcium. Nature 418, 605–611 (2002)
Brown, I. D. & Wu, K. K. Empirical parameters for calculating cation-oxygen bond valences. Acta Crystallogr. B 32, 1957–1959 (1976)
Jewell-Motz, E. A. & Lingrel, J. B. Site-directed mutagenesis of the Na,K-ATPase: consequences of substitutions of negatively-charged amino acids localized in the transmembrane domains. Biochemistry 32, 13523–13530 (1993)
Nielsen, J. M., Pedersen, P. A., Karlish, S. J. & Jorgensen, P. L. Importance of intramembrane carboxylic acids for occlusion of K+ ions at equilibrium in renal Na,K-ATPase. Biochemistry 37, 1961–1968 (1998)
Vilsen, B. & Andersen, J. P. Mutation to the glutamate in the fourth membrane segment of Na+,K+-ATPase and Ca2+-ATPase affects cation binding from both sides of the membrane and destabilizes the occluded enzyme forms. Biochemistry 37, 10961–10971 (1998)
Ogawa, H. & Toyoshima, C. Homology modeling of the cation binding sites of Na+K+-ATPase. Proc. Natl Acad. Sci. USA 99, 15977–15982 (2002)
Ueno, T. & Sekine, T. Study on calcium transport by sarcoplasmic reticulum vesicles using fluorescence probes. J. Biochem. 84, 787–794 (1978)
Pietrobon, D. Familial hemiplegic migraine. Neurotherapeutics 4, 274–284 (2007)
de Carvalho Aguiar, P. et al. Mutations in the Na+/K+ -ATPase α3 gene ATP1A3 are associated with rapid-onset dystonia parkinsonism. Neuron 43, 169–175 (2004)
Cornelius, F., Turner, N. & Christensen, H. R. Modulation of Na,K-ATPase by phospholipids and cholesterol. II. Steady-state and presteady-state kinetics. Biochemistry 42, 8541–8549 (2003)
Sotomayor, C. P., Aguilar, L. F., Cuevas, F. J., Helms, M. K. & Jameson, D. M. Modulation of pig kidney Na+/K+-ATPase activity by cholesterol: role of hydration. Biochemistry 39, 10928–10935 (2000)
Colonna, T. E., Huynh, L. & Fambrough, D. M. Subunit interactions in the Na,K-ATPase explored with the yeast two-hybrid system. J. Biol. Chem. 272, 12366–12372 (1997)
Toyoshima, C. et al. Modeling of the inhibitory interaction of phospholamban with the Ca2+ ATPase. Proc. Natl Acad. Sci. USA 100, 467–472 (2003)
Jones, L. R. Rapid preparation of canine cardiac sarcolemmal vesicles by sucrose flotation. Methods Enzymol. 157, 85–91 (1988)
Skou, J. C. & Esmann, M. Preparation of membrane Na+,K+-ATPase from rectal glands of Squalus acanthias . Methods Enzymol. 156, 43–46 (1988)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Brünger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Murshudov, G. N., Vagin, A. A., Lebedev, A., Wilson, K. S. & Dodson, E. J. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr. D 55, 247–255 (1999)
Laskowski, R. A., Moss, D. S. & Thornton, J. M. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 231, 1049–1067 (1993)
McDonald, I. K. & Thornton, J. M. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 238, 777–793 (1994)
Kraulis, P. J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991)
Merritt, E. A. & Bacon, D. J. Raster3D: Photorealistic molecular graphics. Methods Enzymol. 277, 505–524 (1997)
Acknowledgements
We thank M. Kawamoto and N. Shimizu for their help in data collection at BL41XU, SPring-8, and T. Tsuda for many aspects of this work. We are grateful to D. B. McIntosh for his help in improving the manuscript and H. R.Z. Christensen for technical assistance. Thanks are also due to G. Cramb for sharing sequencing results of the β-subunit with us before publication. This work was supported by a Specially Promoted Project Grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan, to C.T., and grants from the Danish Medical Research Council, to F.C.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Figures S1-S7 with Legends and Supplementary Tables 1 and 2. (PDF 3368 kb)
Rights and permissions
About this article
Cite this article
Shinoda, T., Ogawa, H., Cornelius, F. et al. Crystal structure of the sodium–potassium pump at 2.4 Å resolution. Nature 459, 446–450 (2009). https://doi.org/10.1038/nature07939
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature07939
This article is cited by
-
The effect of cytochrome c on Na,K-ATPase
Journal of Bioenergetics and Biomembranes (2024)
-
The Na+,K+-ATPase and its stoichiometric ratio: some thermodynamic speculations
Biophysical Reviews (2023)
-
Electrostatic switch mechanisms of membrane protein trafficking and regulation
Biophysical Reviews (2023)
-
Identified and potential internalization signals involved in trafficking and regulation of Na+/K+ ATPase activity
Molecular and Cellular Biochemistry (2023)
-
Structure and function of H+/K+ pump mutants reveal Na+/K+ pump mechanisms
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.