Abstract
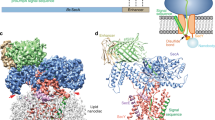
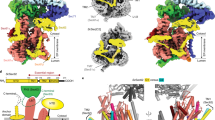
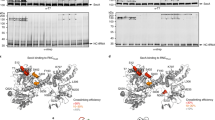
Over 30% of proteins are secreted across or integrated into membranes. Their newly synthesized forms contain either cleavable signal sequences or non-cleavable membrane anchor sequences, which direct them to the evolutionarily conserved Sec translocon (SecYEG in prokaryotes and Sec61, comprising α-, γ- and β-subunits, in eukaryotes). The translocon then functions as a protein-conducting channel1. These processes of protein localization occur either at or after translation. In bacteria, the SecA ATPase2,3 drives post-translational translocation. The only high-resolution structure of a translocon available so far is that for SecYEβ from the archaeon Methanococcus jannaschii4, which lacks SecA. Here we present the 3.2-Å-resolution crystal structure of the SecYE translocon from a SecA-containing organism, Thermus thermophilus. The structure, solved as a complex with an anti-SecY Fab fragment, revealed a ‘pre-open’ state of SecYE, in which several transmembrane helices are shifted, as compared to the previous SecYEβ structure4, to create a hydrophobic crack open to the cytoplasm. Fab and SecA bind to a common site at the tip of the cytoplasmic domain of SecY. Molecular dynamics and disulphide mapping analyses suggest that the pre-open state might represent a SecYE conformational transition that is inducible by SecA binding. Moreover, we identified a SecA–SecYE interface that comprises SecA residues originally buried inside the protein, indicating that both the channel and the motor components of the Sec machinery undergo cooperative conformational changes on formation of the functional complex.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Rapoport, T. A. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450, 663–669 (2007)
Papanikou, E., Karamanou, S. & Economou, A. Bacterial protein secretion through the translocase nanomachine. Nature Rev. Microbiol. 5, 839–851 (2007)
Vrontou, E. & Economou, A. Structure and function of SecA, the preprotein translocase nanomotor. Biochim. Biophys. Acta 1694, 67–80 (2004)
Van den Berg, B. et al. X-ray structure of a protein-conducting channel. Nature 427, 36–44 (2004)
Mori, H. & Ito, K. The Sec protein-translocation pathway. Trends Microbiol. 9, 494–500 (2001)
Cannon, K. S., Or, E., Clemons, W. M., Shibata, Y. & Rapoport, T. A. Disulfide bridge formation between SecY and a translocating polypeptide localizes the translocation pore to the center of SecY. J. Cell Biol. 169, 219–225 (2005)
Tam, P. C., Maillard, A. P., Chan, K. K. & Duong, F. Investigating the SecY plug movement at the SecYEG translocation channel. EMBO J. 24, 3380–3388 (2005)
Li, W. et al. The plug domain of the SecY protein stabilizes the closed state of the translocation channel and maintains a membrane seal. Mol. Cell 26, 511–521 (2007)
Rusch, S. L. & Kendall, D. A. Oligomeric states of the SecA and SecYEG core components of the bacterial Sec translocon. Biochim. Biophys. Acta 1768, 5–12 (2007)
Mitra, K. et al. Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature 438, 318–324 (2005)
Menetret, J. F. et al. Ribosome binding of a single copy of the SecY complex: implications for protein translocation. Mol. Cell 28, 1083–1092 (2007)
Osborne, A. R. & Rapoport, T. A. Protein translocation is mediated by oligomers of the SecY complex with one SecY copy forming the channel. Cell 129, 97–110 (2007)
Kida, Y., Morimoto, F. & Sakaguchi, M. Two translocating hydrophilic segments of a nascent chain span the ER membrane during multispanning protein topogenesis. J. Cell Biol. 179, 1441–1452 (2007)
Douville, K., Price, A., Eichler, J., Economou, A. & Wickner, W. SecYEG and SecA are the stoichiometric components of preprotein translocase. J. Biol. Chem. 270, 20106–20111 (1995)
Lill, R. et al. SecA protein hydrolyzes ATP and is an essential component of the protein translocation ATPase of Escherichia coli . EMBO J. 8, 961–966 (1989)
Mori, H. & Ito, K. Biochemical characterization of a mutationally altered protein translocase: proton motive force stimulation of the initiation phase of translocation. J. Bacteriol. 185, 405–412 (2003)
Mori, H. & Ito, K. An essential amino acid residue in the protein translocation channel revealed by targeted random mutagenesis of SecY. Proc. Natl Acad. Sci. USA 98, 5128–5133 (2001)
Mori, H. & Ito, K. Different modes of SecY-SecA interactions revealed by site-directed in vivo photo-cross-linking. Proc. Natl Acad. Sci. USA 103, 16159–16164 (2006)
Economou, A. & Wickner, W. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell 78, 835–843 (1994)
Vassylyev, D. G. et al. Crystal structure of the translocation ATPase SecA from Thermus thermophilus reveals a parallel, head-to-head dimer. J. Mol. Biol. 364, 248–258 (2006)
Mori, H. et al. Fluorescence resonance energy transfer analysis of protein translocase. SecYE from Thermus thermophilus HB8 forms a constitutive oligomer in membranes. J. Biol. Chem. 278, 14257–14264 (2003)
Breyton, C., Haase, W., Rapoport, T. A., Kuhlbrandt, W. & Collinson, I. Three-dimensional structure of the bacterial protein-translocation complex SecYEG. Nature 418, 662–665 (2002)
Plath, K., Mothes, W., Wilkinson, B. M., Stirling, C. J. & Rapoport, T. A. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell 94, 795–807 (1998)
Sianidis, G. et al. Cross-talk between catalytic and regulatory elements in a DEAD motor domain is essential for SecA function. EMBO J. 20, 961–970 (2001)
Karamanou, S. et al. A molecular switch in SecA protein couples ATP hydrolysis to protein translocation. Mol. Microbiol. 34, 1133–1145 (1999)
Karamanou, S. et al. Preprotein-controlled catalysis in the helicase motor of SecA. EMBO J. 26, 2904–2914 (2007)
Delano, W. L. The PyMOL molecular graphics system. v.0. 97 <http://pymol.sourceforge.net/> (2002)
Kalé, L. et al. NAMD2: Greater scalability for parallel molecular dynamics. J. Comput. Phys. 151, 283–312 (1999)
Shindo, N. et al. Separation of 18 6-aminoquinolyl-carbamyl-amino acids by ion-pair chromatography. Anal. Biochem. 249, 79–82 (1997)
Hunt, J. F. et al. Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science 297, 2018–2026 (2002)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Weeks, C. M. & Miller, R. The design and implementation of SnB v2.0. J. Appl. Crystallogr. 32, 120–124 (1999)
de La Fortelle, E. & Bricogne, G. Maximum-likelihood heavy-atom parameter refinement in the MIR and MAD methods. Methods Enzymol. 276, 472–494 (1997)
Abrahams, J. P. & Leslie, A. G. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D 52, 30–42 (1996)
Cowtan, K. D. & Main, P. Phase combination and cross validation in iterated density-modification calculations. Acta Crystallogr. D 52, 43–48 (1996)
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Brunger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Vagin, A. & Teplyakov, A. MOLREP: an automated program for molecular replacement. in. J. Appl. Crystallogr. 30, 1022–1025 (1997)
Vassylyev, D. G. et al. Structural basis for substrate loading in bacterial RNA polymerase. Nature 448, 163–168 (2007)
Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993)
Vassylyeva, M. N. et al. Cloning, expression, purification, crystallization and initial crystallographic analysis of the preprotein translocation ATPase SecA from Thermus thermophilus. Acta Crystallogr. F 62, 909–912 (2006)
Veenendaal, A. K., van der Does, C. & Driessen, A. J. Mapping the sites of interaction between SecY and SecE by cysteine scanning mutagenesis. J. Biol. Chem. 276, 32559–32566 (2001)
Acknowledgements
We thank K. Inaba, Y. Akiyama and M. Hattori for useful suggestions about sample preparation and crystallization; T. Sakamoto and T. Saika for their assistance in the purification of T. thermophilus SecYE; K. Mochizuki, M. Sano, K. Yoshikaie, T. Adachi and Y. Echizen for technical support; and the beamline staff members at BL41XU of SPring-8 (Sayo, Japan) and NW12 of PF (Tsukuba, Japan) for technical help during data collection. We also thank I. Artsimovitch for critically reading the manuscript. This work was supported by a SORST program grant from JST (Japan Science and Technology) to O.N., by a CREST grant from JST to K.I. and N.D., by a BIRD grant from JST to H.M. and Y.S., by Global COE Program (Center of Education and Research for Advanced Genome-Based Medicine) and a grant for the National Project on Protein Structural and Functional Analyses from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) to O.N., by NIH grants to D.G.V., by grants from MEXT to H.M., S.F., R.I., K.I. and O.N., and by Mitsubishi Foundation grants to O.N.
Author Contributions T.T. carried out the structural determination and the biochemical experiments of T. thermophilus SecYE. H.M. carried out biochemical analyses of SecA–SecY interactions. A.P. and D.G.V. assisted with the crystallization and data collection of SecYE as well as with manuscript preparation. S.F., R.I. and O.N. assisted with the structural determination. T.M. and Y.S. performed the molecular dynamics simulation. N.D. performed disulphide-bond quantification and mass spectrometry. All authors discussed the results and commented on the manuscript. O.N. and K.I. supervised the work and wrote/edited the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary Information
This file contains Supplementary Table 1, Supplementary References, and Supplementary Figures 1-16 with Legends (PDF 17484 kb)
Supplementary Movie 1
This file contains Supplementary Movie 1 (MOV 5693 kb)
Supplementary Movie 2
This file contains Supplementary Movie 2 (MOV 5980 kb)
Rights and permissions
About this article
Cite this article
Tsukazaki, T., Mori, H., Fukai, S. et al. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature 455, 988–991 (2008). https://doi.org/10.1038/nature07421
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature07421
This article is cited by
-
Characterization of the Features of Water Inside the SecY Translocon
The Journal of Membrane Biology (2021)
-
Structure of the substrate-engaged SecA-SecY protein translocation machine
Nature Communications (2019)
-
Structural Basis of the Sec Translocon and YidC Revealed Through X-ray Crystallography
The Protein Journal (2019)
-
Driving Forces of Translocation Through Bacterial Translocon SecYEG
The Journal of Membrane Biology (2018)
-
Two paths diverged in the stroma: targeting to dual SEC translocase systems in chloroplasts
Photosynthesis Research (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.