Abstract
In programmed cell death, a large number of cells undergo apoptosis, and are engulfed by macrophages to avoid the release of noxious materials from the dying cells1,2. In definitive erythropoiesis, nuclei are expelled from erythroid precursor cells and are engulfed by macrophages. Phosphatidylserine is exposed on the surface of apoptotic cells3 and on the nuclei expelled from erythroid precursor cells4; it works as an ‘eat me’ signal for phagocytes5,6. Phosphatidylserine is also expressed on the surface of exosomes involved in intercellular signalling7. Here we established a library of hamster monoclonal antibodies against mouse peritoneal macrophages, and found an antibody that strongly inhibited the phosphatidylserine-dependent engulfment of apoptotic cells. The antigen recognized by the antibody was identified by expression cloning as a type I transmembrane protein called Tim4 (T-cell immunoglobulin- and mucin-domain-containing molecule; also known as Timd4)8. Tim4 was expressed in Mac1+ cells in various mouse tissues, including spleen, lymph nodes and fetal liver. Tim4 bound apoptotic cells by recognizing phosphatidylserine via its immunoglobulin domain. The expression of Tim4 in fibroblasts enhanced their ability to engulf apoptotic cells. When the anti-Tim4 monoclonal antibody was administered into mice, the engulfment of apoptotic cells by thymic macrophages was significantly blocked, and the mice developed autoantibodies. Among the other Tim family members, Tim1, but neither Tim2 nor Tim3, specifically bound phosphatidylserine. Tim1- or Tim4-expressing Ba/F3 B cells were bound by exosomes via phosphatidylserine, and exosomes stimulated the interaction between Tim1 and Tim4. These results indicate that Tim4 and Tim1 are phosphatidylserine receptors for the engulfment of apoptotic cells, and may also be involved in intercellular signalling in which exosomes are involved.
Similar content being viewed by others
Main
Caspase-activated DNase (CAD)-deficient cells do not undergo apoptotic DNA fragmentation, but their DNA is degraded in phagocytes after they are engulfed9. We used this knowledge to assay the engulfment of apoptotic cells, and identified MFG-E8 (milk fat globule EGF factor 8) that stimulates their engulfment10. MFG-E8 is expressed in thioglycollate-elicited peritoneal macrophages, tingible-body macrophages of lymph nodes and spleen, Langerhans cells and mammary epithelial cells10,11,12,13. Hu et al.14 reported that naïve peritoneal macrophages engulf apoptotic cells in a phosphatidylserine (PS)-dependent manner. We confirmed this with our assay system, which uses CAD-/- thymocytes as prey (Fig. 1a). Peritoneal macrophages efficiently engulfed apoptotic cells, and this was inhibited by the D89E mutant of MFG-E8 that masks PS10. However, peritoneal macrophages expressed little MFG-E8 (see below), and this MFG-E8 deficiency did not affect the ability of these macrophages to engulf apoptotic cells (data not shown). To identify the molecules involved in this engulfment, Armenian hamsters were immunized with mouse peritoneal cells, and hybridomas were prepared. Among 1,200 hybridomas, one monoclonal antibody (Kat5-18) dose-dependently inhibited the phagocytosis of apoptotic cells by peritoneal macrophages (Fig. 1a). Most Mac1+ peritoneal cells were stained by Kat5-18 (Fig. 1b), indicating that peritoneal macrophages expressed its antigen.
a, Mouse peritoneal cells were used for phagocytosis with D89E10 or Kat5-18, stained for TUNEL, analysed by flow cytometry, and the percentage of TUNEL+ macrophages determined. Average values of two experiments are shown. b, Peritoneal macrophages incubated with biotin-Kat5-18, followed with FITC-streptavidin and APC-anti-Mac1. c, Parental and Tim4-expressing Ba/F3 analysed by flow cytometry with Kat5-18. d, mRNA levels of Tim4 and MFG-E8 in resident or thioglycollate-elicited peritoneal macrophages, and bone marrow-derived immature dendritic cells, determined by real-time PCR. e, Tim4 mRNA levels in mouse tissues, determined by real-time PCR. f, Real-time PCR for Tim4 mRNA in Mac1+ and Mac1- cells from lymph nodes, spleen and fetal liver.
To identify the antigen for Kat5-18, we used retrovirus-mediated expression cloning15. A complementary DNA library was constructed with messenger RNA from mouse peritoneal cells, converted to retroviruses, and used to infect mouse Ba/F3 cells. After 2 days in culture, transformants labelled by Kat5-18 (less than 0.1%) were sorted using a cell sorter, expanded in culture, and sorted again. This cycle of sorting and expansion was repeated three times until most cells stained with Kat5-18. The sorted Kat5-18+ cells carried 2–3 different integrated cDNAs, and Tim4 cDNA was found in two independent experiments. When Tim4 cDNA was introduced into Ba/F3, the transformants stained strongly with Kat5-18 (Fig. 1c), confirming that Tim4 was the antigen recognized by Kat5-18. Real-time PCR indicated that Tim4 mRNA was expressed in resident, but not thioglycollate-elicited, peritoneal cells (Fig. 1d), in sharp contrast with MFG-E8, which was expressed in thioglycollate-elicited but weakly in resident peritoneal cells. A similar situation was observed with bone marrow-derived immature dendritic cells, which expressed a high level of MFG-E8, but not Tim4. Other major tissues that expressed Tim4 included the spleen, thymus, lymph nodes and salivary glands (Fig. 1e). When cells from the spleen, lymph nodes and fetal liver were sorted into Mac1+ and Mac1- cells using MACS (magnetic cell sorting), Tim4 mRNA was found only in Mac1+ cells (Fig. 1f). Flow cytometry indicated that about 20% of Mac1+ cells in the lymph nodes expressed Tim4 (Supplementary Fig. 1).
DNase II degrades DNA of apoptotic cells in lysosomes after phagocytes engulf them16. To examine the role of Tim4 in the engulfment of apoptotic cells, a fibroblast cell line expressing Tim4 was established with NIH3T3 expressing DNase II10 (NIH3T3/DNase II/Tim4) (Supplementary Fig. 2). The ability of the parental NIH3T3/DNase II to engulf apoptotic cells was weak: when incubated with apoptotic CAD-/- thymocytes for 120 min, only 20% of the cells engulfed them (Fig. 2a). On the other hand, more than 50% of NIH3T3/DNase II/Tim4 engulfed apoptotic cells within 60 min. This effect of Tim4 was confirmed by microscopic observation. That is, when apoptotic cells were cultured with NIH3T3/DNase II, few apoptotic cells were seen attached to NIH3T3/DNase II (Fig. 2b). In contrast, many apoptotic cells associated with NIH3T3/DNase II/Tim4, and many were TUNEL-positive, indicating that these cells had been engulfed. NIH3T3/DNase II/Tim4 engulfed apoptotic cells but not healthy ones, and this engulfment was dose-dependently inhibited by Kat5-18 (Fig. 2c), confirming that Tim4 conferred the ability to engulf apoptotic cells on NIH3T3/DNase II. To confirm the function of Tim4 in vivo, CAD-/- mice were given Kat5-18, and treated with dexamethasone to induce apoptosis in the thymus. As shown in Fig. 2d, in the thymus of mice that received the control IgG, 32.3% of F4/80+ macrophages carried more than 15 TUNEL+ apoptotic cells. This percentage of the macrophages carrying more than 15 apoptotic cells was reduced to about 12.6% in the thymus of the mice that received Kat5-18. Inefficient engulfment of apoptotic cells often leads to autoimmune diseases17. In fact, the mice that received Kat5-18 developed autoantibodies (anti-cardiolipin and anti-double-stranded-DNA antibodies) within 5 weeks in their serum (Fig. 2e). Two mice strongly responded to Kat5-18 and two weakly, but the injection of normal IgG or PBS never caused the development of autoantibodies. The serum anti-cardiolipin antibody level was reduced when the injection of Kat5-18 was interrupted, confirming that this increase was due to Kat5-18.
a, NIH3T3/DNase II (filled squares, open squares) and NIH3T3/DNase II/Tim4 (filled circles, open circles) incubated with 3.0 × 105 (filled squares, filled circles) or 6.0 × 105 (open squares, open circles) apoptotic CAD-/- thymocytes for the indicated periods. The percentage of TUNEL+ macrophages was determined by flow cytometry. Average values from three experiments are shown; error bars, ±1s.e. b, NIH3T3/DNase II and NIH3T3/DNase II/Tim4 used for 60-min phagocytosis, stained for TUNEL, and observed by microscopy. Arrows indicate TUNEL+ cells. c, NIH3T3/DNase II or NIH3T3/DNase II/Tim4 incubated with healthy or apoptotic thymocytes in the presence of Kat5-18, stained for TUNEL. The percentage of TUNEL+ macrophages was determined by flow cytometry. Average values from three experiments are shown; error bars, ±1s.e.. d, CAD-/- mice that had received normal hamster Ab or Kat5-18 were treated with dexamethasone. Top, the thymus stained for TUNEL (green) and F4/80 (red). The number of TUNEL+ cells determined for 50 randomly selected macrophages (bottom). Experiments were carried out three times, and the percentage of the macrophages carrying more than 15 apoptotic cells plotted (right). Mann–Whitney’s U-test was used to analyse the difference, and P values are shown. e, Normal hamster IgG, Kat5-18 or PBS was injected into mice twice a week for 5 weeks. The serum concentration of anti-cardiolipin and anti-dsDNA was determined at 2, 5 and 10 weeks. P values determined as above are shown. A492, absorbance at 492 nm.
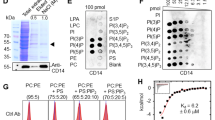
Mouse Tim4 is a type I membrane protein, consisting of a signal sequence and extracellular, transmembrane and cytosolic regions (Fig. 3a). To examine how Tim4 enhances the engulfment of apoptotic cells, the Tim4 extracellular region was fused to the Fc region of human IgG (Tim4-Fc). The purified Tim4-Fc showed a band at 82 kDa in SDS–PAGE under reducing conditions, and a 170 kDa band under non-reducing conditions (Supplementary Fig. 3), indicating that Tim4-Fc existed as a dimer. When Ba/F3 expressing Fas was treated with Fas ligand, they underwent apoptosis and bound annexin V (Fig. 3b), which binds to PS exposed on the surface of apoptotic cells18. These cells also bound Tim4-Fc, with a time course similar to their binding of annexin V. When a nitrocellulose filter spotted with various phospholipids was incubated with Tim4-Fc, Tim4-Fc bound PS, but not phosphatidylcholine, phosphatidylinositol, or phosphatidylethanolamine (Fig. 3c). The affinity of Tim4-Fc for PS, determined with microtitre plates coated with phospholipids, was comparable to that of MFG-E8 (Kd ≈ 2 nM)(Fig. 3d). These results suggested that Tim4 recognizes PS exposed on apoptotic cells as an engulfment signal. In support of this idea, the MFG-E8 mutant D89E dose-dependently inhibited the engulfment of apoptotic cells by NIH3T3/DNase II/Tim4 (Supplementary Fig. 4).
a, Structure of Tim4, showing signal sequence (S), IgV-like (IgV), mucin-like (Mucin), transmembrane (TM) and cytoplasmic regions (Cyt). a.a., amino acid. Tim4-Fc carries the Tim4 extracellular region and the Fc region of human IgG. b, Ba/F3-Fas treated with Fas ligand for the indicated periods, stained with annexin V or Tim4-Fc. c, A filter spotted with phosphatidylcholine (PC), phosphatidylinositol (PI), phosphatidylethanolamine (PE) or phosphatidylserine (PS), incubated with Tim4-Fc followed by anti-IgG. d, Microtitre plates coated with indicated phospholipids incubated with Tim4-Fc, followed by anti-IgG. The assay was performed in triplicate. e, Lipid overlay assay performed as c with Tim1-Fc, Tim2-Fc and Tim3-Fc. f, Structures of the extracellular region of Tim1, Tim2 and Tim4, and their hybrids, fused to human IgG. g, Microtitre plates coated with PS incubated with increasing concentrations of Tim1-Fc, Tim2-Fc, Tim4-Fc, or their hybrids.
The Tim family has three members in human (TIM1, 3 and 4), and eight in mouse (Tim1–8). To examine whether other Tim family members can recognize PS, the Fc fusion proteins were prepared with mouse Tim1, Tim2 and Tim3 (Supplementary Fig. 5). Tim1-Fc, Tim2-Fc and Tim3-Fc each behaved as a dimer (Supplementary Fig. 6). The lipid overlay assay indicated that Tim1-Fc, but neither Tim2-Fc nor Tim3-Fc, specifically bound PS (Fig. 3e). Accordingly, full-length Tim1, but neither Tim2 nor Tim3, enhanced the engulfment of apoptotic cells when it was expressed in NIH3T3/DNase II cells (Supplementary Fig. 7). The extracellular region of Tim family members consists of two domains (IgG V-chain like (IgV) and mucin domains)8. To examine which domain of Tim1 and Tim4 is responsible for binding PS, IgV and mucin domains were swapped between Tim1 and Tim2, and between Tim4 and Tim2, and fused to the Fc region (Fig. 3f). As shown in Fig. 3g, the chimaeric molecules carrying IgV domain from Tim1 or Tim4 tightly bound PS, while no binding was observed with the chimaeric molecules carrying mucin domain from Tim1 or Tim4.
Various mammalian cells produce exosomes that expose PS on their surface7. The ability of Tim1 and Tim4 to bind PS suggested that exosomes bind to cells expressing Tim1 or Tim4. To examine this possibility, Ba/F3 cell lines expressing Tim1 or Tim4 (Ba/F3-Tim1 or Ba/F3-Tim4) were established. The growing Ba/F3-Tim1 or Ba/F3-Tim4 but not the parental Ba/F3 bound annexin V (Fig. 4a, and data not shown). Ba/F3-Tim1 and Ba/F3-Tim4 were then treated with biotin-labelled annexin V, followed by staining with gold-conjugated streptavidin. Observation by electron microscopy indicated that Ba/F3-Tim1 or Ba/F3-Tim4 was associated with exosome-like vesicles, and several gold particles were found on the outer leaflet of the exosomes (Fig. 4b). Such vesicles were not observed with the Ba/F3 parental cells. Meyers et al.19 reported that Tim4 acts as a ligand for Tim1 expressed in Th2 cells. Accordingly, Tim1-Fc strongly bound Ba/F3-Tim4 (Fig. 4c). In addition, Tim1-Fc bound Ba/F3-Tim1, and Tim4-Fc bound Ba/F3-Tim4. The ability of Tim4 and Tim1 to bind PS suggested that these homophilic and heterophilic interactions between Tim1 and Tim4 can be stimulated by exosomes. In support of this idea, the binding of Tim4-Fc to Ba/F3-Tim4 was inhibited by annexin V (Fig. 4d). To directly confirm the effect of exosomes on association between Tim1 and Tim4, microspheres were coated with Tim4-Fc, and subjected to binding assay with Tim1-Fc. As shown in Fig. 4e, little Tim1-Fc bound to Tim4-Fc-coated beads, and exosomes from Ba/F3 dose-dependently stimulated the binding of Tim1-Fc to the beads.
a, Ba/F3-Tim4 incubated with PE-annexin V, and analysed by flow cytometry. Staining profile of Ba/F3 in red. b, Electron micrographs of Ba/F3, Ba/F3-Tim1 or Ba/F3-Tim4, incubated with biotin-annexin V and stained with streptavidin-colloidal gold. Gold particles on exosome-like vesicles indicated by arrowheads. Scale bar, 0.3 μm. c, Ba/F3-Tim4 or Ba/F3-Tim1 incubated with Tim1-Fc or Tim4-Fc, stained with FITC-anti-IgG, and analysed by flow cytometry. Staining profiles of Ba/F3 in red. d, Ba/F3-Tim4 incubated with Tim4-Fc in the absence or presence of annexin V, stained with FITC-anti-IgG. e, Tim4-Fc-coated latex beads incubated with Tim1-Fc in the absence (green) or presence of 10-fold (blue) or 50-fold (brown) concentrated exosomes, stained with PE-anti-Tim1, and analysed by flow cytometry. Profile without Tim1-Fc in red dotted line.
Phosphatidylserine exposed on the outer leaflet of the plasma membrane is often used as a recognition signal for phagocytes to engulf apoptotic cells. Fadok et al. reported that phosphatidylserine receptor (PSR), a ubiquitously expressed type II-membrane protein, binds to PS on apoptotic cells and mediates their engulfment20, and initial experiments with PSR-/- mice supported this notion21,22. However, a careful analysis of PSR-/- mice by Bose et al.23 indicated that PSR is not involved in the engulfment of apoptotic cells, and it is unlikely to work as a receptor for PS. Here, we showed that Tim4 expressed in macrophages in various tissues mediates the engulfment of apoptotic cells by recognizing PS, indicating that Tim4 is the phosphatidylserine receptor for engulfment of apoptotic cells. Tim1 also enhanced the PS-dependent engulfment of apoptotic cells. Tim1 is expressed in kidney tubule cells post-ischaemia24, suggesting that it may have a role in removing injured and dead cells, to help restore the morphological integrity of the kidney. It will be interesting to examine whether any other molecules work as phosphatidylserine receptors.
Some non-apoptotic T or B cells25,26 as well as macrophages27 have been reported to bind annexin V. Here, we showed that the cells expressing Tim1 or Tim4 associated with exosomes that carried exposed PS. Activated Th2 cells and B cells express Tim128,29, while the resident macrophages express Tim4, suggesting that the non-apoptotic annexin V-binding cells are those that express Tim1 or Tim4. Exosomes are secreted from various cells and act as an intercellular signalling device7. Tim1 and Tim4 may mediate this signalling by enhancing the uptake of exosomes. Cells expressing Tim1 or Tim4 tended to aggregate (M.M. and S.N., unpublished results). PS on activated T cells is suggested to function in cell-to-cell contact at the immunological synapse25. Thus, Tim1 and Tim4 may also be involved in cell-to-cell interactions via PS on exosomes. Finally, Tim family genes of human and mouse are located on a genetic interval that has linkage to a number of autoimmune diseases, such as asthma, allergy and atopy30. The establishment of Tim1 and Tim4 as PS receptors may contribute to the understanding of these autoimmune diseases.
Methods Summary
Production of monoclonal antibodies
Armenian hamsters (Oriental Yeast) were immunized with mouse peritoneal cells, and about 1,200 hybridomas were established by fusing cells from the popliteal and inguinal lymph nodes with NSObcl-2 mouse myeloma. The hybridoma supernatants were tested by a phagocytosis assay, and one hybridoma (Kat5-18) that inhibited the engulfment of apoptotic cells was identified.
Construction of cDNA library, and expression cloning
The double-stranded cDNA longer than 1.0 kb was ligated into a BstXI-digested pMXs vector15. Escherichia coli DH10B was transformed by electroporation, and about 1.0 × 106 clones were produced. Plasmid DNA from the cDNA library was introduced into PLAT-E packaging cells, and the culture supernatant was used to infect Ba/F3. A subpopulation of Ba/F3 that stained intensely with Kat5-18 was selected by repeated FACS. The integrated cDNA was recovered by PCR with the vector primers from the genomic DNA of Ba/F3 transformants.
Phagocytosis assay
To assay the engulfment of apoptotic cells, CAD-/- thymocytes were induced to undergo apoptosis with leucine-zipper tagged Fas ligand, and co-cultured with peritoneal macrophages or NIH3T3/DNase II cells. The cells were fixed with paraformaldehyde, subjected to TUNEL staining, and analysed by flow cytometry using a FACSAria (BD Biosciences).
References
Danial, N. N. & Korsmeyer, S. J. Cell death: critical control points. Cell 116, 205–219 (2004)
Savill, J., Dransfield, I., Gregory, C. & Haslett, C. A blast from the past: clearance of apoptotic cells regulates immune responses. Nature Rev. Immunol. 2, 965–975 (2002)
Fadok, V. A. et al. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J. Immunol. 148, 2207–2216 (1992)
Yoshida, H. et al. Phosphatidylserine-dependent engulfment by macrophages of nuclei from erythroid precursor cells. Nature 437, 754–758 (2005)
Tanaka, Y. & Schroit, A. J. Insertion of fluorescent phosphatidylserine into the plasma membrane of red blood cells. Recognition by autologous macrophages. J. Biol. Chem. 258, 11335–11343 (1983)
Schlegel, R. A. & Williamson, P. Phosphatidylserine, a death knell. Cell Death Differ. 8, 551–563 (2001)
Thery, C., Zitvogel, L. & Amigorena, S. Exosomes: composition, biogenesis and function. Nature Rev. Immunol. 2, 569–579 (2002)
Kuchroo, V. K., Umetsu, D. T., DeKruyff, R. H. & Freeman, G. J. The TIM gene family: emerging roles in immunity and disease. Nature Rev. Immunol. 3, 454–462 (2003)
Nagata, S. DNA degradation in development and programmed cell death. Annu. Rev. Immunol. 23, 853–875 (2005)
Hanayama, R. et al. Identification of a factor that links apoptotic cells to phagocytes. Nature 417, 182–187 (2002)
Hanayama, R. & Nagata, S. Impaired involution of mammary glands in the absence of milk fat globule EGF factor 8. Proc. Natl Acad. Sci. USA 102, 16886–16891 (2005)
Hanayama, R. et al. Autoimmune disease and impaired uptake of apoptotic cells in MFG-E8-deficient mice. Science 304, 1147–1150 (2004)
Miyasaka, K., Hanayama, R., Tanaka, M. & Nagata, S. Expression of milk fat globule epidermal growth factor 8 in immature dendritic cells for engulfment of apoptotic cells. Eur. J. Immunol. 34, 1414–1422 (2004)
Hu, B., Sonstein, J., Christensen, P. J., Punturieri, A. & Curtis, J. L. Deficient in vitro and in vivo phagocytosis of apoptotic T cells by resident murine alveolar macrophages. J. Immunol. 165, 2124–2133 (2000)
Kitamura, T. et al. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp. Hematol. 31, 1007–1014 (2003)
Kawane, K. et al. Impaired thymic development in mouse embryos deficient in apoptotic DNA degradation. Nature Immunol. 4, 138–144 (2003)
Gaipl, U. S. et al. Impaired clearance of dying cells in systemic lupus erythematosus. Autoimmun. Rev. 4, 189–194 (2005)
Koopman, G. et al. Annexin V for flow cytometric detection of phosphatidylserine expression on B cells undergoing apoptosis. Blood 84, 1415–1420 (1994)
Meyers, J. H. et al. TIM-4 is the ligand for TIM-1, and the TIM-1-TIM-4 interaction regulates T cell proliferation. Nature Immunol. 6, 455–464 (2005)
Fadok, V. A. et al. A receptor for phosphatidylserine-specific clearance of apoptotic cells. Nature 405, 85–90 (2000)
Li, M. O., Sarkisian, M. R., Mehal, W. Z., Rakic, P. & Flavell, R. A. Phosphatidylserine receptor is required for clearance of apoptotic cells. Science 302, 1560–1563 (2003)
Kunisaki, Y. et al. Defective fetal liver erythropoiesis and T lymphopoiesis in mice lacking the phosphatidylserine receptor. Blood 103, 3362–3364 (2004)
Bose, J. et al. The phosphatidylserine receptor has essential functions during embryogenesis but not in apoptotic cell removal. J. Biol. 3, 15 (2004)
Ichimura, T. et al. Kidney injury molecule-1 (KIM-1), a putative epithelial cell adhesion molecule containing a novel immunoglobulin domain, is up-regulated in renal cells after injury. J. Biol. Chem. 273, 4135–4142 (1998)
Fischer, K. et al. Antigen recognition induces phosphatidylserine exposure on the cell surface of human CD8+ T cells. Blood 108, 4094–4101 (2006)
Dillon, S. R., Mancini, M., Rosen, A. & Schlissel, M. S. Annexin V binds to viable B cells and colocalizes with a marker of lipid rafts upon B cell receptor activation. J. Immunol. 164, 1322–1332 (2000)
Callahan, M. K., Williamson, P. & Schlegel, R. A. Surface expression of phosphatidylserine on macrophages is required for phagocytosis of apoptotic thymocytes. Cell Death Differ. 7, 645–653 (2000)
Umetsu, S. E. et al. TIM-1 induces T cell activation and inhibits the development of peripheral tolerance. Nature Immunol. 6, 447–454 (2005)
Gielen, A. W. et al. Expression of T cell immunoglobulin- and mucin-domain-containing molecules-1 and -3 (TIM-1 and -3) in the rat nervous and immune systems. J. Neuroimmunol. 164, 93–104 (2005)
McIntire, J. J. et al. Identification of Tapr (an airway hyperreactivity regulatory locus) and the linked Tim gene family. Nature Immunol. 2, 1109–1116 (2001)
Acknowledgements
We thank R. Hanayama for advice in the initial stage of this project, and M. Fujii and M. Harayama for secretarial assistance. This work was supported in part by Grants-in-Aid from the Ministry of Education, Science, Sports, and Culture in Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Methods with additional references and Supplementary Figures 1-7 with Legends. (PDF 1430 kb)
Rights and permissions
About this article
Cite this article
Miyanishi, M., Tada, K., Koike, M. et al. Identification of Tim4 as a phosphatidylserine receptor. Nature 450, 435–439 (2007). https://doi.org/10.1038/nature06307
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature06307
This article is cited by
-
Exosome-based delivery strategies for tumor therapy: an update on modification, loading, and clinical application
Journal of Nanobiotechnology (2024)
-
Apoptotic bodies: bioactive treasure left behind by the dying cells with robust diagnostic and therapeutic application potentials
Journal of Nanobiotechnology (2023)
-
Adipose tissue macrophages: implications for obesity-associated cancer
Military Medical Research (2023)
-
Proteogenomic links to human metabolic diseases
Nature Metabolism (2023)
-
Internalization of apoptotic cells during efferocytosis requires Mertk-mediated calcium influx
Cell Death & Disease (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.