Abstract
The mitotic spindle is typically thought of as an array of microtubules, microtubule-associated proteins and motors that self-organizes to align and segregate chromosomes1. The major spindle components consist of proteins and DNA, the primary structural elements of the spindle1. Other macromolecules including RNA and lipids also associate with spindles, but their spindle function, if any, is unknown. Poly(ADP-ribose) (PAR) is a large, branched, negatively charged polymeric macromolecule whose polymerization onto acceptor proteins is catalysed by a family of poly(ADP-ribose) polymerases (PARPs)2. Several PARPs localize to the spindle in vertebrate cells, suggesting that PARPs and/or PAR have a role in spindle function2. Here we show that PAR is enriched in the spindle and is required for spindle function—PAR hydrolysis or perturbation leads to rapid disruption of spindle structure, and hydrolysis during spindle assembly blocks the formation of bipolar spindles. PAR exhibits localization dynamics that differ from known spindle proteins and are consistent with a low rate of turnover in the spindle. Thus, PAR is a non-proteinaceous, non-chromosomal component of the spindle required for bipolar spindle assembly and function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Wittmann, T., Hyman, A. & Desai, A. The spindle: a dynamic assembly of microtubules and motors. Nature Cell Biol. 3, E28–E34 (2001)
Smith, S. The world according to PARP. Trends Biochem. Sci. 26, 174–179 (2001)
Tulin, A., Stewart, D. & Spradling, A. C. The Drosophila heterochromatic gene encoding poly(ADP-ribose) polymerase (PARP) is required to modulate chromatin structure during development. Genes Dev. 16, 2108–2119 (2002)
Hatakeyama, K., Nemoto, Y., Ueda, K. & Hayaishi, O. Purification and characterization of poly(ADP-ribose) glycohydrolase. Different modes of action on large and small poly(ADP-ribose). J. Biol. Chem. 261, 14902–14911 (1986)
Saxena, A., Saffery, R., Wong, L. H., Kalitsis, P. & Choo, K. H. Centromere proteins Cenpa, Cenpb, and Bub3 interact with poly(ADP-ribose) polymerase-1 protein and are poly(ADP-ribosyl)ated. J. Biol. Chem. 277, 26921–26926 (2002)
Kickhoefer, V. A. et al. The 193-kD vault protein, VPARP, is a novel poly(ADP-ribose) polymerase. J. Cell Biol. 146, 917–928 (1999)
Smith, S. & de Lange, T. Tankyrase promotes telomere elongation in human cells. Curr. Biol. 10, 1299–1302 (2000)
Sbodio, J. I. & Chi, N. W. Identification of a tankyrase-binding motif shared by IRAP, TAB182, and human TRF1 but not mouse TRF1. NuMA contains this RXXPDG motif and is a novel tankyrase partner. J. Biol. Chem. 277, 31887–31892 (2002)
Bakondi, E. et al. Detection of poly(ADP-ribose) polymerase activation in oxidatively stressed cells and tissues using biotinylated NAD substrate. J. Histochem. Cytochem. 50, 91–98 (2002)
Earle, E. et al. Poly(ADP-ribose) polymerase at active centromeres and neocentromeres at metaphase. Hum. Mol. Genet. 9, 187–194 (2000)
Smith, S. & de Lange, T. Cell cycle dependent localization of the telomeric PARP, tankyrase, to nuclear pore complexes and centrosomes. J. Cell Sci. 112, 3649–3656 (1999)
Sawin, K. E. & Mitchison, T. J. Mitotic spindle assembly by two different pathways in vitro. J. Cell Biol. 112, 925–940 (1991)
Slama, J. T. et al. Specific inhibition of poly(ADP-ribose) glycohydrolase by adenosine diphosphate (hydroxymethyl)pyrrolidinediol. J. Med. Chem. 38, 389–393 (1995)
Wilde, A. & Zheng, Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science 284, 1359–1362 (1999)
Sawin, K. E., LeGuellec, K., Philippe, M. & Mitchison, T. J. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature 359, 540–543 (1992)
Compton, D. A., Szilak, I. & Cleveland, D. W. Primary structure of NuMA, an intranuclear protein that defines a novel pathway for segregation of proteins at mitosis. J. Cell Biol. 116, 1395–1408 (1992)
Dionne, M. A., Howard, L. & Compton, D. A. NuMA is a component of an insoluble matrix at mitotic spindle poles. Cell Motil. Cytoskel. 42, 189–203 (1999)
Olmsted, J. B., Stemple, D. L., Saxton, W. M., Neighbors, B. W. & McIntosh, J. R. Cell cycle-dependent changes in the dynamics of MAP2 and MAP4 in cultured cells. J. Cell Biol. 109, 211–223 (1989)
Kapoor, T. M. & Mitchison, T. J. Eg5 is static in bipolar spindles relative to tubulin: evidence for a static spindle matrix. J. Cell Biol. 154, 1125–1133 (2001)
Saxton, W. M. et al. Tubulin dynamics in cultured mammalian cells. J. Cell Biol. 99, 2175–2186 (1984)
Wittmann, T., Wilm, M., Karsenti, E. & Vernos, I. TPX2, A novel Xenopus MAP involved in spindle pole organization. J. Cell Biol. 149, 1405–1418 (2000)
Poirier, G. G., de Murcia, G., Jongstra-Bilen, J., Niedergang, C. & Mandel, P. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc. Natl Acad. Sci. USA 79, 3423–3427 (1982)
Aoufouchi, S. & Shall, S. Regulation by phosphorylation of Xenopus laevis poly(ADP-ribose) polymerase enzyme activity during oocyte maturation. Biochem. J. 325, 543–551 (1997)
Chi, N. W. & Lodish, H. F. Tankyrase is a golgi-associated mitogen-activated protein kinase substrate that interacts with IRAP in GLUT4 vesicles. J. Biol. Chem. 275, 38437–38444 (2000)
Rouleau, M., Aubin, R. A. & Poirier, G. G. Poly(ADP-ribosyl)ated chromatin domains: access granted. J. Cell Sci. 117, 815–825 (2004)
Pickett-Heaps, J. D., Forer, A. & Spurck, T. Traction fibre: toward a “tensegral” model of the spindle. Cell Motil. Cytoskel. 37, 1–6 (1997)
Troll, W., Garte, S. & Frenkel, K. Suppression of tumor promotion by inhibitors of poly(ADP)ribose formation. Basic Life Sci. 52, 225–232 (1990)
Tirnauer, J. S., Salmon, E. D. & Mitchison, T. J. Microtubule plus-end dynamics in Xenopus egg extract spindles. Mol. Biol. Cell 15, 1776–1784 (2004)
Lin, W., Ame, J. C., Aboul-Ela, N., Jacobson, E. L. & Jacobson, M. K. Isolation and characterization of the cDNA encoding bovine poly(ADP-ribose) glycohydrolase. J. Biol. Chem. 272, 11895–11901 (1997)
Acknowledgements
We thank A. Pollington, B. Ward, J. Tirnauer and B. Brieher for critical reading of the manuscript; A. Straight for CENPE antibodies; A. Groen and D. Miyamoto for TPX2, NuMA and Eg5 antibodies; Z. Perlman for assistance with data analysis; D. L. Coyle for technical support; and the Nikon Imaging Facility at Harvard Medical School for use of microscopes. This work was supported by NIH grants to T.M.J. and M.K.J.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Supplementary Figure S1
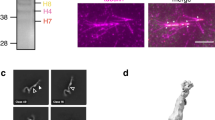
PAR Localization to the kinetochore. Xenopus extract spindles were incubated with Alexa 488-labeled PAR antibody, and X-rhodamine-labeled CENPE antibody (CENP) and, visualized using spinning disk confocal microscopy every 30 s for 20 min. PAR co-localized with CENPE at every time point. In Merge, PAR is green and CENPE, red. (JPG 36 kb)
Supplementary Movie M1
Real time spinning disk confocal microscopy of Xenopus egg extract spindles treated with 100 µg/ml PARG. Images were obtained every 30s for 15 min. (MP4 156 kb)
Supplementary Movie M2
Real time spinning disk confocal microscopy of Xenopus egg extract spindles treated with 500 µg/ml anti PAR antibody. Images were obtained every 30s for 15 min. (MP4 186 kb)
Rights and permissions
About this article
Cite this article
Chang, P., Jacobson, M. & Mitchison, T. Poly(ADP-ribose) is required for spindle assembly and structure. Nature 432, 645–649 (2004). https://doi.org/10.1038/nature03061
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature03061
This article is cited by
-
GmPARPs differentially regulate the drought and heat stress tolerance in soybean
Plant Growth Regulation (2023)
-
Miki (Mitotic Kinetics Regulator) Immunoexpression in Normal Liver, Cirrhotic Areas and Hepatocellular Carcinomas: a Preliminary Study with Clinical Relevance
Pathology & Oncology Research (2020)
-
Structural analyses of NudT16–ADP-ribose complexes direct rational design of mutants with improved processing of poly(ADP-ribosyl)ated proteins
Scientific Reports (2019)
-
PARylation regulates stress granule dynamics, phase separation, and neurotoxicity of disease-related RNA-binding proteins
Cell Research (2019)
-
Poly(ADP-ribose) mediates asymmetric division of mouse oocyte
Cell Research (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.