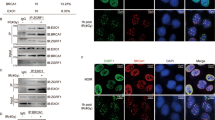
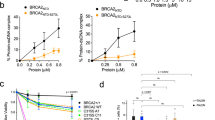
Abstract
The breakpoint cluster region of the MLL gene (MLLbcr) is frequently rearranged in therapy-related and infant acute leukaemia, but the destabilizing mechanism is poorly understood. We recently proposed that DNA replication stress results in MLLbcr cleavage via endonuclease G (EndoG) and represents the common denominator of genotoxic therapy-induced MLL destabilization. Here we performed a siRNA screen for new factors involved in replication stress-induced MLL rearrangements employing an enhanced green fluorescent protein-based reporter system. We identified 10 factors acting in line with EndoG in MLLbcr breakage or further downstream in the repair of the MLLbcr breaks, including activation-induced cytidine deaminase (AID), previously proposed to initiate MLLbcr rearrangements in an RNA transcription-dependent mechanism. Further analysis connected AID and EndoG in MLLbcr destabilization via base excision repair (BER) components. We show that replication stress-induced recruitment of EndoG to the MLLbcr and cleavage are AID/BER dependent. Notably, inhibition of the core BER factor Apurinic-apyrimidinic endonuclease 1 protects against MLLbcr cleavage in tumour and human cord blood-derived haematopoietic stem/progenitor cells, harbouring the cells of origin of leukaemia. We propose that off-target binding of AID to the MLLbcr initiates BER-mediated single-stranded DNA cleavage, which causes derailed EndoG activity ultimately resulting in leukaemogenic MLLbcr rearrangements.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Gole B, Wiesmüller L . Leukemogenic rearrangements at the mixed lineage leukemia gene (MLL)- multiple rather than a single mechanism. Front Cell Dev Biol 2015; 3: 41.
Shivakumar R, Tan W, Wilding GE, Wang ES, Wetzler M . Biologic features and treatment outcome of secondary acute lymphoblastic leukemia–a review of 101 cases. Ann Oncol 2008; 19: 1634–1638.
Stanulla M, Wang J, Chervinsky DS, Thandla S, Aplan PD . DNA cleavage within the MLL breakpoint cluster region is a specific event which occurs as part of higher-order chromatin fragmentation during the initial stages of apoptosis. Mol Cell Biol 1997; 17: 4070–4079.
Sim SP, Liu LF . Nucleolytic cleavage of the mixed lineage leukemia breakpoint cluster region during apoptosis. J Biol Chem 2001; 276: 31590–31595.
Broeker PL, Super HG, Thirman MJ, Pomykala H, Yonebayashi Y, Tanabe S et al. Distribution of 11q23 breakpoints within the MLL breakpoint cluster region in de novo acute leukemia and in treatment-related acute myeloid leukemia: correlation with scaffold attachment regions and topoisomerase II consensus binding sites. Blood 1996; 87: 1912–1922.
Hensel JP, Gillert E, Fey GH, Marschalek R . Breakpoints of t(4;11) translocations in the human MLL and AF4 genes in ALL patients are preferentially clustered outside of high-affinity matrix attachment regions. J Cell Biochem 2001; 82: 299–309.
Strissel PL, Strick R, Rowley JD, Zeleznik-Le NJ . An in vivo topoisomerase II cleavage site and a DNase I hypersensitive site colocalize near exon 9 in the MLL breakpoint cluster region. Blood 1998; 92: 3793–3803.
Hars ES, Lyu YL, Lin CP, Liu LF . Role of apoptotic nuclease caspase-activated DNase in etoposide-induced treatment-related acute myelogenous leukemia. Cancer Res 2006; 66: 8975–8979.
Liu X, He Y, Li F, Huang Q, Kato TA, Hall RP et al. Caspase-3 promotes genetic instability and carcinogenesis. Mol Cell 2015; 58: 284–296.
Gole B, Baumann C, Mian E, Ireno CI, Wiesmüller L . Endonuclease G initiates DNA rearrangements at the MLL breakpoint cluster upon replication stress. Oncogene 2015; 34: 3391–3401.
Scharf S, Zech J, Bursen A, Schraets D, Oliver PL, Kliem S et al. Transcription linked to recombination: a gene-internal promoter coincides with the recombination hot spot II of the human MLL gene. Oncogene 2007; 26: 1361–1371.
Meng FL, Du Z, Federation A, Hu J, Wang Q, Kieffer-Kwon KR et al. Convergent transcription at intragenic super-enhancers targets AID-initiated genomic instability. Cell 2014; 159: 1538–1548.
Wright RL, Slemmons KK, Vaughan AT . Estradiol induces gene proximity and MLL-MLLT3 fusion in an activation-induced cytidine deaminase-mediated pathway. Leuk Lymphoma 2015; 56: 1460–1465.
Keimling M, Deniz M, Varga D, Stahl A, Schrezenmeier H, Kreienberg R et al. The power of DNA double-strand break (DSB) repair testing to predict breast cancer susceptibility. FASEB J 2012; 26: 2094–2104.
Kraft D, Rall M, Volcic M, Metzler E, Groo A, Stahl A et al. NF-κB-dependent DNA damage-signaling differentially regulates DNA double-strand break repair mechanisms in immature and mature human hematopoietic cells. Leukemia 2015; 29: 1543–1554.
Akyüz N, Boehden GS, Süsse S, Rimek A, Preuss U, Scheidtmann KH et al. DNA substrate dependence of p53-mediated regulation of double-strand break repair. Mol Cell Biol 2002; 22: 6306–6317.
Ireno IC, Baumann C, Stöber R, Hengstler JG, Wiesmüller L . Fluorescence-based recombination assay for sensitive and specific detection of genotoxic carcinogens in human cells. Arch Toxicol 2014; 88: 1141–1159.
Biechonski S, Gourevich D, Rall M, Aqaqe N, Yassin M, Zipin-Roitman A et al. Quercetin alters the DNA damage response in human hematopoietic stem and progenitor cells via TopoII- and PI3K-dependent mechanisms synergizing in leukemogenic rearrangements. Int J Cancer 2017; 140: 864–876.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13: 2498–2504.
Dohrn L, Salles D, Siehler SY, Kaufmann J, Wiesmüller L . BRCA1-mediated repression of mutagenic end-joining of DNA double-strand breaks requires complex formation with BACH1. Biochem J 2012; 441: 919–926.
Hampp S, Kiessling T, Buechle K, Mansilla SF, Thomale J, Rall M et al. DNA damage tolerance pathway involving DNA polymerase ι and the tumor suppressor p53 regulates DNA replication fork progression. Proc Natl Acad Sci USA 2016; 113: E4311–E4319.
Carey MF, Peterson CL, Smale ST . Transcriptional Regulation in Eukaryotes: Concepts, Strategies and Techniques, 2nd edn. Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2009.
Mirault ME, Boucher P, Tremblay A . Nucleotide-resolution mapping of topoisomerase-mediated and apoptotic DNA strand scissions at or near an MLL translocation hotspot. Am J Hum Genet 2006; 79: 779–791.
Wang Z, Huang Y, Zhang J . Molecularly targeting the PI3K-Akt-mTOR pathway can sensitize cancer cells to radiotherapy and chemotherapy. Cell Mol Biol Lett 2014; 19: 233–242.
Chafin DR, Guo H, Price DH . Action of alpha-amanitin during pyrophosphorolysis and elongation by RNA polymerase II. J Biol Chem 1995; 270: 19114–19119.
Lange SS, Takata K, Wood RD . DNA polymerases and cancer. Nat Rev Cancer 2011; 11: 96–110.
Vaidyanathan B, Yen WF, Pucella JN, Chaudhuri J . AIDing chromatin and transcription-coupled orchestration of immunoglobulin class-switch recombination. Front Immunol 2014; 5: 120.
Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM . DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 1998; 273: 5858–5868.
Kalinowska M, Garncarz W, Pietrowska M, Garrard WT, Widlak P . Regulation of the human apoptotic DNase/RNase endonuclease G: involvement of Hsp70 and ATP. Apoptosis 2005; 10: 821–830.
Vallur AC, Maizels N . Activities of human exonuclease 1 that promote cleavage of transcribed immunoglobulin switch regions. Proc Natl Acad Sci USA 2008; 105: 16508–16512.
Horton JK, Prasad R, Hou E, Wilson SH . Protection against methylation-induced cytotoxicity by DNA polymerase beta-dependent long patch base excision repair. J Biol Chem 2000; 275: 2211–2218.
Liu T, Huang J . DNA end resection: facts and mechanisms. Genomics proteomics. Bioinformatics 2016; 14: 126–130.
Zhao GY, Sonoda E, Barber LJ, Oka H, Murakawa Y, Yamada K et al. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol Cell 2007; 25: 663–675.
Beck C, Boehler C, Guirouilh Barbat J, Bonnet ME, Illuzzi G, Ronde P et al. PARP3 affects the relative contribution of homologous recombination and nonhomologous end-joining pathways. Nucleic Acids Res 2014; 42: 5616–5632.
Singh DK, Karmakar P, Aamann M, Schurman SH, May A, Croteau DL et al. The involvement of human RECQL4 in DNA double-strand break repair. Aging Cell 2010; 9: 358–371.
Santarpia L, Iwamoto T, Di Leo A, Hayashi N, Bottai G, Stampfer M et al. DNA repair gene patterns as prognostic and predictive factors in molecular breast cancer subtypes. Oncologist 2013; 18: 1063–1073.
Shamanna RA, Singh DK, Lu H, Mirey G, Keijzers G, Salles B et al. RECQ helicase RECQL4 participates in non-homologous end joining and interacts with the Ku complex. Carcinogenesis 2014; 35: 2415–2424.
Guo F, Li J, Du W, Zhang S, O'Connor M, Thomas G et al. mTOR regulates DNA damage response through NF-κB-mediated FANCD2 pathway in hematopoietic cells. Leukemia 2013; 27: 2040–2046.
Choi EJ, Cho BJ, Lee DJ, Hwang YH, Chun SH, Kim HH et al. Enhanced cytotoxic effect of radiation and temozolomide in malignant glioma cells: targeting PI3K-AKT-mTOR signaling, HSP90 and histone deacetylases. BMC Cancer 2014; 14: 17.
Kumar A, Fernandez-Capetillo O, Carrera AC . Nuclear phosphoinositide 3-kinase beta controls double-strand break DNA repair. Proc Natl Acad Sci USA 2010; 107: 7491–7496.
Groth A, Corpet A, Cook AJ, Roche D, Bartek J, Lukas J et al. Regulation of replication fork progression through histone supply and demand. Science 2007; 318: 1928–1931.
Schatz DG, Swanson PC . V(D)J recombination: mechanisms of initiation. Annu Rev Genet 2011; 45: 167–202.
Geng LY, Shi ZZ, Dong Q, Cai XH, Zhang YM, Cao W et al. Expression of SNC73, a transcript of the immunoglobulin alpha-1 gene, in human epithelial carcinomas. World J Gastroenterol 2007; 13: 2305–2311.
Papaemmanuil E, Rapado I, Li Y, Potter NE, Wedge DC, Tubio J et al. RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia. Nat Genet 2014; 46: 116–125.
Tsai AG, Lu H, Raghavan SC, Muschen M, Hsieh CL, Lieber MR . Human chromosomal translocations at CpG sites and a theoretical basis for their lineage and stage specificity. Cell 2008; 135: 1130–1142.
Pauklin S, Sernández IV, Bachmann G, Ramiro AR, Petersen-Mahrt SK . Estrogen directly activates AID transcription and function. J Exp Med 2009; 206: 99–111.
Sawai Y, Kodama Y, Shimizu T, Ota Y, Maruno T, Eso Y et al. Activation-induced cytidine deaminase contributes to pancreatic tumorigenesis by inducing tumor-related gene mutations. Cancer Res 2015; 75: 3292–3301.
Matsumoto T, Shimizu T, Nishijima N, Ikeda A, Eso Y, Matsumoto Y et al. Hepatic inflammation facilitates transcription-associated mutagenesis via AID activity and enhances liver tumorigenesis. Carcinogenesis 2015; 36: 904–913.
Hasham MG, Snow KJ, Donghia NM, Branca JA, Lessard MD, Stavnezer J et al. Activation-induced cytidine deaminase-initiated off-target DNA breaks are detected and resolved during S phase. J Immunol 2012; 189: 2374–2382.
Watanabe N, Wachi S, Fujita T . Identification and characterization of BCL-3-binding protein: implications for transcription and DNA repair or recombination. J Biol Chem 2003; 278: 26102–26110.
Macheret M, Halazonetis TD . DNA replication stress as a hallmark of cancer. Annu Rev Pathol 2015; 10: 425–448.
Delker RK, Zhou Y, Strikoudis A, Stebbins CE, Papavasiliou FN . Solubility-based genetic screen identifies RING finger protein 126 as an E3 ligase for activation-induced cytidine deaminase. Proc Natl Acad Sci USA 2013; 110: 1029–1034.
Fu D, Calvo JA, Samson LD . Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat Rev Cancer 2012; 12: 104–120.
Cortizas EM, Zahn A, Safavi S, Reed JA, Vega F, Di Noia JM et al. UNG protects B cells from AID-induced telomere loss. J Exp Med 2016; 213: 2459–2472.
Balakrishnan L, Bambara RA . Flap endonuclease 1. Annu Rev Biochem 2013; 82: 119–138.
Wang G, Vasquez KM . Impact of alternative DNA structures on DNA damage, DNA repair, and genetic instability. DNA Repair (Amst) 2014; 19: 143–151.
Maga G, van Loon B, Crespan E, Villani G, Hübscher U . The block of DNA polymerase delta strand displacement activity by an abasic site can be rescued by the concerted action of DNA polymerase beta and Flap endonuclease 1. J Biol Chem 2009; 284: 14267–14275.
Eid W, Steger M, El-Shemerly M, Ferretti LP, Peña-Diaz J, König C et al. DNA end resection by CtIP and exonuclease 1 prevents genomic instability. EMBO Rep 2010; 11: 962–968.
Strick R, Strissel PL, Borgers S, Smith SL, Rowley JD . Dietary bioflavonoids induce cleavage in the MLL gene and may contribute to infant leukemia. Proc Natl Acad Sci USA 2000; 97: 4790–4795.
Walter D, Lier A, Geiselhart A, Thalheimer FB, Huntscha S, Sobotta MC et al. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature 2015; 520: 549–552.
Acknowledgements
We thank Daniela Waldraff, Regina Häfele and Jasmin Petermeier (all from the Department of Obstetrics and Gynaecology, Ulm University, Ulm, Germany) for their help with experimental procedures. This work was financially supported by the German Research Foundation (DFG, WI3099/7-1, WI3099/7-2 and PA3 in Research Training Group 1789 ‘Cellular and Molecular Mechanisms in aging’, CEMMA) and the German Space Agency/German Ministry of Education and Research (DLR/BMBF, project TRANSCAN ERA-NET 01KT1302).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Rights and permissions
About this article
Cite this article
Gole, B., Mian, E., Rall, M. et al. Base excision repair proteins couple activation-induced cytidine deaminase and endonuclease G during replication stress-induced MLL destabilization. Leukemia 32, 159–167 (2018). https://doi.org/10.1038/leu.2017.191
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2017.191